Using Graph Neural Networks, Explainable AI, Property Graphs & Virtual Reality for computational support in discovering and developing drugs, vaccines & therapies
The Covid-19 pandemic has shown us that discovering & developing novel drugs and vaccines in much shorter times is essential – and achievable, given that we had several vaccines developed, tested and available within the first year of the pandemic. The research behind these vaccines were a good deal older, of course, but there is one additional element that helped accelerate the whole process, the hidden hero that doesn´t always get credited for the contribution: Computational tools like molecular modeling & simulation tools, which are predicted to cut the cost and the time to develop new treatments with as much as 30% by 2030.
This is not really a surprise, but the biomedical world is an experimental/empirical world that did not adopt computational tools and modeling techniques as early as the engineering disciplines and other professions. That has been changing rapidly due to the recent challenges where tools & techniques like modeling, simulation, AI & machine learning have started proving their worth in biomedical research & industry but simulating the dynamic nature of molecules is highly complex, still requiring powerful computing resource, efficient data structures and algorithms.
We will be looking at four interesting challenges, each potentially more than one MS theses, with the last one being a more practical theme. All themes are related, making it possible to define a thesis subject across these themes, and co-operating with other master´s students within these themes. In any case, the thesis subject will need to be discussed and scoped correctly so get in touch if you are interested in any one of the following themes:
Theme 1: Efficient representation of molecular structures
Graph data structures – especially property graphs – have already been pointed out as natural models for the representation of molecular structures because they resemble graphs and can be parametrized to represent the characteristics of each atom in a molecule. See figure below.
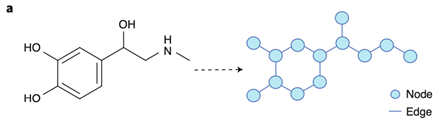
[Ref. figure 3a in Jiménez-Luna et al.1]
Still, current representations are static and do not consider the dynamic nature of the chemical bonds between the atoms that can be stronger or weaker, or may disappear and re-appear, etc. The thesis will investigate existing solutions that use property graphs, propose alternative property graph models/representations of chemical and biological structures and benchmark them primarily in terms of their storage and performance characteristics. One may also wish to look at the limitations of property graphs and propose extensions to property graphs (which we call extended property graphs or EPGs, somewhat like what Junghanns et al. proposed in 20162) to enable better modeling of dynamic molecular structures.
See also a very recent paper by Davit et al.3 that offers a summary of molecular representations in AI-driven drug discovery, which is relevant for both this theme and the next.
Theme 2: Graph Neural Networks for learning to recognize and match molecular structures
As in many other disciplines, artificial intelligence and machine learning have started to have a central importance also in biomedical sciences & technologies, especially in discovering & developing drugs, including vaccines and therapeutics. Discovering a drug is a demanding exercise, which can be summarized as matching certain structural and physicochemical properties of a drug-candidate molecule to a target molecule (the virus spike protein, some molecule causing a disorder, maybe a hormone, etc.). One computational AI/ML technique that is already being used for structural/visual matching of properties & patterns is the convolutional neural network (CNN), which is a deep learning technique. See the following Wikipedia article that offers more information: https://en.wikipedia.org/wiki/Convolutional_neural_network.
Graphs with properties are popular and effective data structures for representing molecular structures and their properties as pointed out in theme 1 and the figure above. In this master´s thesis, we will be looking at a generalized version of the CNN called Graph Neural networks (GNN). The ultimate goal is to create & benchmark a learning algorithm (or several) capable of recognizing patterns/properties that match a drug candidate to a drug target, which is a step towards automating drug discovery4.
Theme 3: Explainable AI
Drug development can take up to 15-20 years, and it can be divided into 2 major phases:
- pre-clinical studies where the drug is discovered and tested in a wet lab and on animals (rat etc.)
- clinical studies where the drug is tested on human.
Before being tested on humans, the therapeutic molecules must be approved by the administration authorities. For the approval, information about practically all steps of the drug development process will need to be provided. When deep learning techniques are used in any one of these steps (in discovery, and especially in testing & drug efficacy calculations, risk assessments etc.), the information that needs to be provided to the authorities may not be obvious due to the “black-box” nature of regular neural networks.
This master´s thesis theme will focus upon Explainable Artificial Intelligence (XAI1,5) to develop and benchmark deep learning algorithms that also provide transparency of the operation, justification of the results, information for supporting the decisions of the authorities and for estimating uncertainty and potential deviations/risks in the results.
Theme 4: Molecular Dynamics in Virtual Reality
There are several software tools that offer visualization of molecular structures and for facilitating drug discovery. One common tool, also used in biosciences and chemistry education at the University of Oslo and in many other educational institutions is PyMOL, an open-source Python software for molecular visualization (see https://pymol.org/2/).
There are also more comprehensive and specialized systems for drug discovery, but most of them have interfaces designed for experts who understand not only biology and chemistry but also computational biomedicine, which is a very specialized field. This hinders many regular professionals of biomedicine and chemistry form using these tools without expert help. In addition, most tools offer 2D or 3D visualization on a 2D plane (the screen), and very few tools offer the possibility to interact with molecular structures and manipulate them directly.
It has been shown that 3D Virtual Reality has the potential to offer very intuitive, immersive and interactive user interfaces. There have been some experimental attempts to move visualization to VR and to add some amount of manipulation capabilities (see for example Prof. David Glowacki´s work here: https://glow-wacky.com/).
This master thesis will aim to create a VR user interface and platform for existing open-source software for molecular visualization. This is a more practical thesis subject that will require programming skills in addition to an understanding of the interaction between the front-end rendering client and the back-end high-performance computing architecture for the compute-intensive simulations. It will most likely also require some understanding of the effectiveness of the underlying data structures that represent the molecules, which may imply cooperation with the master students working on theme 1.
--
Note again that these themes need to be discussed and scoped correctly for a master´s thesis. Please get in touch with one of us if you are interested and let us have a talk.
Bibliography
1. Jiménez-Luna J, Grisoni F, Schneider G. Drug discovery with explainable artificial intelligence. Nature Machine Intelligence. 2020;2(10):573-584. doi:10.1038/s42256-020-00236-4
2. Junghanns M, Petermann A, Teichmann N, Gómez K, Rahm E. Analyzing Extended Property Graphs with Apache Flink. Published online 2016. doi:10.1145/2980523.2980527
3. David L, Thakkar A, Mercado R, Engkvist O. Molecular representations in AI-driven drug discovery: a review and practical guide. Journal of Cheminformatics. 2020;12(1):1-22. doi:10.1186/s13321-020-00460-5
4. Xiong J, Xiong Z, Chen K, Jiang H, Zheng M. Graph neural networks for automated de novo drug design. Drug discovery today. Published online February 17, 2021. doi:10.1016/j.drudis.2021.02.011
5. Jiménez-Luna J, Skalic M, Weskamp N, Schneider G. Coloring Molecules with Explainable Artificial Intelligence for Preclinical Relevance Assessment. Journal of Chemical Information and Modeling. 2021;61(3):1083-1094. doi:10.1021/acs.jcim.0c01344
