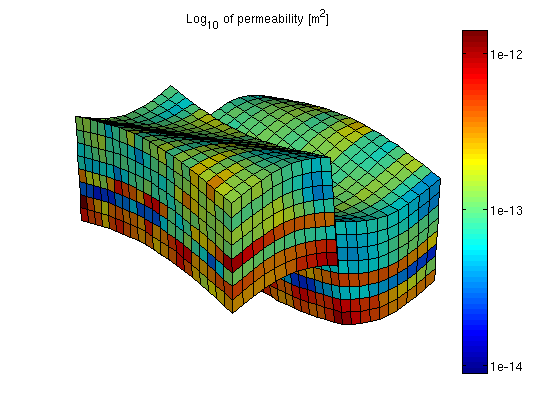
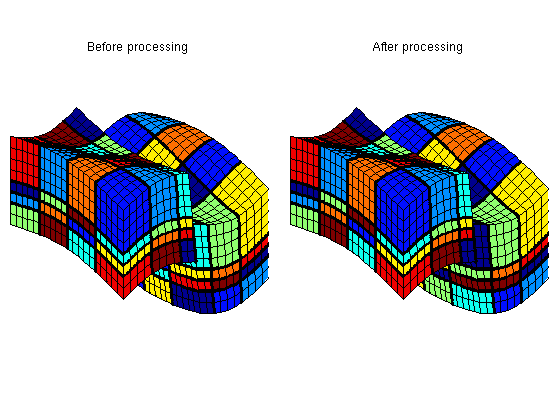
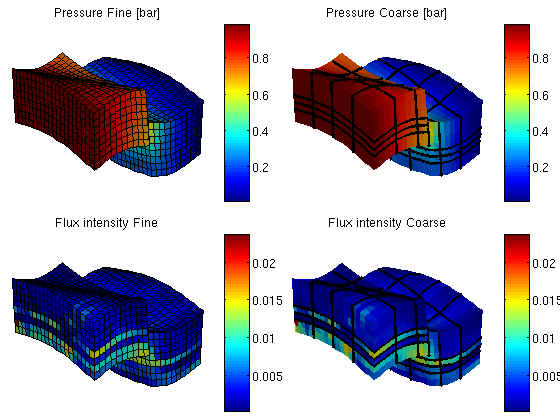
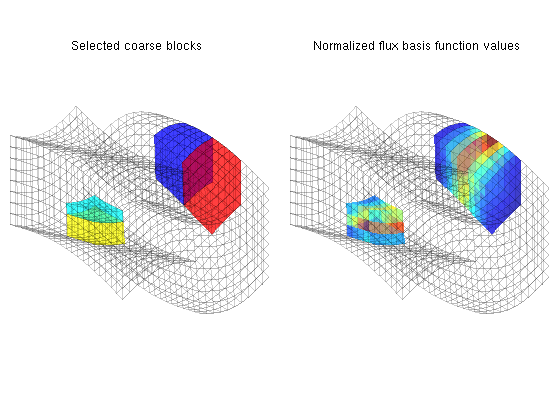
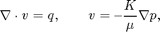You are here:
MRST
/
Modules
/
Multiscale Mixed Finite Elements Module
/
Multiscale Flow Solver: II