GoTools Parametrization Library
1.1
This module is part of the SINTEF GoTools libraries and licenced under the GNU General Public License
The purpose of the GoTools Parametrization Library is to compute a reasonable parametrization for a discrete geometrical object. This geometrical object can be a planar graph embedded in 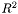 or
or 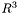 (triangulation, rectangular grid, etc.) or a point cloud without any explicit connectivity, but with an identified border.
(triangulation, rectangular grid, etc.) or a point cloud without any explicit connectivity, but with an identified border.
There are many contexts for which such a parametrization is useful, for example when mapping textures onto polygonal meshes or when triangulating a surface from a set of scattered 3D points. The algorithms contained in this software are based on the work of Dr. Michael Floater.
This library has three degrees of freedom for specifying the parametrization:
-
choice of data structure
-
choice of boundary parametrization
-
choice of parametrization of the interior
As mentioned in the last section, the data to be parametrized can be of various kinds, from general polygonal meshes (3D points and connectivity) to point clouds (no connectivity information, but boundary nodes must be indentified and ordered). The various possible data types are given as sub-classes of PrOrganizedPoints. You should choose the one that corresponds to the data that you want to parametrize. Many of these classes have read/write functionality for a generic format; you might want to reimplement these to correspond with the format of the data you are using. The list of currently available data structures are:
You can of course write a totally new specialization of the PrOrganizedPoints class, as long as you implement all the required member functions.
The boundary of the surface must be parametrized before the surface interior. This is done by the class PrParametrizeBdy. There are several ways of parametrizing the boundary. You can specify the method you want to use through the PrParametrizeBdy::setParamKind() member function. There are currently three options:
-
PrCHORDLENGTHBDY - parametrize by chordlength (generally preferred)
-
PrCENTRIPETAL - parametrize using square root of boundary point distances
-
PrUNIFBDY - parametrize by imposing a fixed distance between each boundary point
Another thing that must be specified when determining the parametrization of the boundary is the shape of the parametrical domain. It can be a circle (default), or a rectangle (corners must be specified).
The parametrization of the interior can also be done in several ways, which are each implemented as a sub-class of PrParametrizeInt. Currently available methods are: (click on the links for individual explanations)
The general procedure for parametrizing a data set can be summarized as follows:
- Prepare the data structure
- Parametrize the boundary
- Establish a PrParametrizeBdy object.
- Associate it with your data by using the PrParametrizeBdy::attach() function.
- Choose parametrization type by calling the PrParametrizeBdy::setParamKind() function.
- Execute the parametrization by calling PrParametrizeBdy(). When calling this function without arguments, the parametrical domain will be a circle. A rectangular parameter domain can also be used. This is specified by additional arguments to the function.
- Parametrize the interior
- Accessing the result
- The parametrization can be accessed in several ways, through the PrOrganizedPoints interface of your data structure:
- Most of the sub-classes of PrOrganizedPoints also have print() and scan() functions that can be used for reading and writing their data to a stream.
- You can write your own customised data access and streaming functions.
The procedure mentioned above can be observed in a working program located in the app/ folder. The source file name is demo.C. The program is run from the command line, where you can also specify the data. It can handle both point clouds and triangulations. A sample point cloud and triangulation is provided in the data/ folder.
For instance, try
/// app/demo -i data/noh-mask.tri
///
/// app/demo -i data/noh-mask.pcloud
///
If the input data is a triangulation (first case above), the resulting, parametrized triangulation will be saved in the file triangulation. In both cases, the parametrization for each node/point will be dumped to the file uv_nodes.
Good luck!
Generated on Tue Jun 12 11:05:05 2007 for GoTools Parametrization Library by
 1.5.1
1.5.1
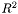 or
or 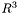 (triangulation, rectangular grid, etc.) or a point cloud without any explicit connectivity, but with an identified border.
(triangulation, rectangular grid, etc.) or a point cloud without any explicit connectivity, but with an identified border. 1.5.1
1.5.1