ad-core: Automatic Differentiation Core¶
Object-oriented framework for solvers based on automatic differentiation (MRST AD-OO). This module by itself does not contain any complete simulators, but rather implements the common framework used for many other modules. For instance, the ad-blackoil module contains models for black-oil equations that inherit all basic features from ad-core, the ad-eor module inherits models from ad-blackoil, and so on.
Overview of the solvers:
Models¶
Base classes¶
-
Contents¶ MODELS
- Files
- ExtendedReservoirModel - PhysicalModel - Base class for all AD models. Implements a generic discretized model. ReservoirModel - Base class for physical models WrapperModel - Wrapper model which can be inherited for operations on the parent
-
class
PhysicalModel(G, varargin)¶ Base class for all AD models. Implements a generic discretized model.
Synopsis:
model = PhysicalModel(G)
Description:
Base class for implementing physical models for use with automatic differentiation. This class cannot be used directly.
A physical model consists of a set of discrete operators that can be used to define the model equations and a nonlinear tolerance that defines how close the values must be to zero before the equations can be considered to be fulfilled. In most cases, the operators are defined over a grid, which is an optional property in this class. In addition, the class contains a flag informing if the model equations are linear, and a flag determining verbosity of class functions.
The class contains member functions for:
- evaluating residual equations and Jacobians
- querying and setting individual variables in the physical state
- executing a single nonlinear step (i.e., a linear solve with a possible subsequent stabilization step), verifying convergence, and reporting the status of the step
- verifying the model, associated physical states, or individual physical properties
as well as a number of utility functions for updating the physical state with increments from the linear solve, etc. See the implementation of the class for more details.
Parameters: G – Simulation grid. Can be set to empty. Keyword Arguments: ‘property’ – Set property to the specified value. Returns: model – Class instance of PhysicalModel.Note
This is the standard base class for the AD-OO solvers. As such, it does not implement any specific discretization or equations and is seldom instansiated on its own.
See also
ReservoirModel,ThreePhaseBlackOilModel,TwoPhaseOilWaterModel-
capProperty(model, state, name, minvalue, maxvalue)¶ Ensure that a property is within a specific range by capping.
Synopsis:
state = model.capProperty(state, 'someProp', minValOfProp) state = model.capProperty(state, 'someProp', minValOfProp, maxValOfProp)
Parameters: - model – Class instance.
- state – State `struct`to be updated.
- name – Name of the field to update, as supported by
model.getVariableField. - minvalue – Minimum value of state property
name. Any values below this threshold will be set to this value. Set to-inffor no lower bound. - maxvalue – OPTIONAL: Maximum value of state property
name. Values that are larger than this limit are set to the limit. For no upper limit, setinf.
Returns: state – State
structwherenameis within the limits.Example:
% Make a random field, and limit it to the range [0.2, 0.5]. state = struct('pressure', rand(10, 1)) state = model.capProperty(state, 0.2, 0.5); disp(model.getProp(state, 'pressure'))
-
checkConvergence(model, problem, varargin)¶ Check and report convergence based on residual tolerances
Synopsis:
[convergence, values, names] = model.checkConvergence(problem)
Description:
Basic convergence testing for a linearized problem. By default, this simply takes the inf norm of all model equations. Subclasses are free to overload this function for more sophisticated and robust options.
Parameters: - model – Class instance
- problem –
LinearizedProblemADto be checked for convergence. The default behavior is to check all equations againstmodel.nonlinearTolerancein the inf/max norm. - n – OPTIONAL· The norm to be used. Default:
inf.
Returns: - convergence – Vector of length
Nwith bools indicatingtrue/falseif that residual/error measure has converged. - values – Vector of length
Ncontaining the numerical values checked for convergence. - names – Cell array of length
Ncontaining the names tested for convergence.
Note
By default,
Nis equal to the number of equations inproblemand the convergence simply checks the convergence of each equation against a genericnonlinearTolerance. However, subclasses are free to produce any number of convergence criterions and they need not correspond to specific equations at all.
-
checkProperty(model, state, property, n_el, dim)¶ Check dimensions of property and throw error if dims do not match
Synopsis:
model.checkProperty(state, 'pressure', G.cells.num, 1); model.checkProperty(state, 'components', [ncell, ncomp], [1, 2]);
Parameters: - model – Class instance.
- state – State `struct`to be checked.
- name – Name of the field to check, as supported by
model.getVariableField. - n_el – Array of length
Nwhere entryi`corresponds to the size of the property in dimension `dim(i). - dim – Array of length
Ncorresponding to the dimensions for whichn_elis to be checked.
-
getAdjointEquations(model, state0, state, dt, forces, varargin)¶ Get the adjoint equations (please read note before use!)
Synopsis:
[problem, state] = model.getAdjointEquations(state0, state, dt, drivingForces)
Description:
Function to get equation when using adjoint to calculate gradients. This make it possible to use different equations to calculate the solution in the forward mode, for example if equations are solved explicitly as for hysteretic models. it is assumed that the solution of the system in forward for the two different equations are equal i.e
problem.val == 0.Parameters: - model – Class instance
- state – Current state to be solved for time t + dt.
- state0 – The converged state at time t.
- dt – The scalar time-step.
- forces – Forces struct. See
getDrivingForces.
Returns: - problem –
LinearizedProblemADderived class containing the linearized equations used for the adjoint problem. This function is normallygetEquationsand assumes that the function supports thereverseModeargument. - state – State. Possibly updated. See
getEquationsfor details.
Keyword Arguments: ‘reverseMode’ – If set to true, the reverse mode of the equations are provided.
Note
A caveat is that this function provides the forward-mode version of the adjoint equations, normally identical to
getEquations. MRST allows for separate implementations of adjoint and regular equations in order to allow for rigorous treatment of hysteresis and other semi-explicit parameters.
-
getDrivingForces(model, control)¶ Get driving forces in expanded format.
Synopsis:
vararg = model.getDrivingForces(schedule.control(ix))
Parameters: - model – Class instance
- control – Struct with the driving forces as fields. This should
be a struct with the same fields as in
getValidDrivingForces, although this is not explicitly enforced in this routine.
Returns: vararg –
Cell-array of forces in the format:
{'W', W, 'bc', bc, ...}
This is typically used as input to variable input argument functions that support various boundary conditions options.
-
getEquations(model, state0, state, dt, forces, varargin)¶ Get the set of linearized model equations with possible Jacobians
Synopsis:
[problem, state] = model.getEquations(state0, state, dt, drivingForces)
Description:
Provide a set of linearized equations. Unless otherwise noted, these equations will have
ADItype, containing both the value and Jacobians of the residual equations.Parameters: - model – Class instance
- state – Current state to be solved for time t + dt.
- state0 – The converged state at time t.
- dt – The scalar time-step.
- forces – Forces struct. See
getDrivingForces.
Keyword Arguments: - ‘resOnly’ – If supported by the equations, this flag will result in only the values of the equations being computed, omitting any Jacobians.
- ‘iteration’ – The nonlinear iteration number. This can be provided so that the underlying equations can account for the progress of the nonlinear solution process in a limited degree, for example by updating some quantities only at the first iteration.
Returns: - problem – Instance of the wrapper class
LinearizedProblemADcontaining the residual equations as well as other useful information. - state – The equations are allowed to modify the system state, allowing a limited caching of expensive calculations only performed when necessary.
See also
getAdjointEquations
-
getIncrement(model, dx, problem, name)¶ Get specific named increment from a list of different increments.
Synopsis:
dv = model.getIncrement(dx, problem, 'name')
Description:
Find increment in linearized problem with given name, or output zero if not found. A linearized problem can give updates to multiple variables and this makes it easier to get those values without having to know the order they were input into the constructor.
Parameters: - model – Class instance.
- dx – Cell array of increments corresponding to the names
in
problem.primaryVariables. - problem – Instance of
LinearizedProblemfrom which the increments were computed. - name – Name of the variable for which the increment is to be extracted.
Returns: dv – The value of the increment, if it is found. Otherwise a scalar zero value is returned.
-
getMaximumTimestep(model, state, state0, dt, drivingForces)¶ Define the maximum allowable time-step based on physics or discretization choice
-
getModelEquations(model, state0, state, dt, drivingForces)¶ Get equations from AD states
-
getPrimaryVariables(model, state)¶ Get primary variables from state
-
getProp(model, state, name)¶ Get a single property from the nonlinear state
Synopsis:
p = model.getProp(state, 'pressure');
Parameters: - model – Class instance.
- state –
structholding the state of the nonlinear problem. - name – A property name supported by the model’s
getVariableFieldmapping.
Returns: - p – Property taken from the state.
- state – The state (modified if property evaluation was done)
See also
getProps
-
getProps(model, state, varargin)¶ Get multiple properties from state in one go
Synopsis:
[p, s] = model.getProps(state, 'pressure', 's');
Parameters: - model – Class instance.
- state –
structholding the state of the nonlinear problem.
Keyword Arguments: ‘FieldName’ – Property names to be extracted. Any number of properties can be requested.
Returns: varargout – Equal number of output arguments to the number of property strings sent in, corresponding to the respective properties.
See also
getProp
-
getValidDrivingForces(model)¶ Get a struct with the default valid driving forces for the model
Synopsis:
forces = model.getValidDrivingForces();
Description:
Different models support different types of boundary conditions. Each model should implement a default struct, showing the solvers what a typical allowed struct of boundary conditions looks like in terms of the named fields present.
Parameters: model – Class instance Returns: forces – A struct with any number of fields. The fields must be present, but they can be empty.
-
getVariableField(model, name, throwError)¶ Map known variable by name to field and column index in
stateSynopsis:
[fn, index] = model.getVariableField('someKnownField')
Description:
Get the index/name mapping for the model (such as where pressure or water saturation is located in state). For this parent class, this always result in an error, as this model knows of no variables.
For subclasses, however, this function is the primary method the class uses to map named values (such as the name of a component, or the human readable name of some property) and the compact representation in the state itself.
Parameters: - model – Class instance.
- name – The name of the property for which the storage field in
stateis requested. Attempts at retrieving a field the model does not know results in an error. - throwError – OPTIONAL: If set to false, no error is thrown and empty fields are returned.
Returns: - fn – Field name in the
structwherenameis stored. - index – Column index of the data.
See also
getProp,setProp
-
incrementProp(model, state, name, increment)¶ Increment named state property by given value
Synopsis:
state = model.incrementProp(state, 'PropertyName', increment)
Parameters: - model – Class instance.
- state –
structholding the state of the nonlinear problem. - name – Name of the property to updated. See
getVariableField - value – The increment that will be added to the current value
of property
name.
Returns: state – Updated state
struct.Example:
% For a model which knows of the field 'pressure', increment % the value by 7 so that the final value is 10 (=3+7). state = struct(‘pressure’, 3); state = model.incrementProp(state, ‘pressure’, 7);
-
initStateAD(model, state, vars, names, origin)¶ Initialize AD state from double state
-
static
limitUpdateAbsolute(dv, maxAbsCh)¶ Limit a update by absolute limit
-
static
limitUpdateRelative(dv, val, maxRelCh)¶ Limit a update by relative limit
-
makeStepReport(model, varargin)¶ Get the standardized report all models provide from
stepFunctionSynopsis:
report = model.makeStepReport('Converged', true);
Description:
Normalized struct with a number of useful fields. The most important fields are the fields representing
FailureandConvergedwhichNonLinearSolverreacts appropriately to.Parameters: model – Class instance Keyword Arguments: various – Keyword/value pairs that override the default values. Returns: report – Normalized report with defaulted values where not provided. See also
stepFunction,NonLinearSolverAD
-
prepareReportstep(model, state, state0, dT, drivingForces)¶ Prepare state and model (temporarily) before solving a report-step
-
prepareTimestep(model, state, state0, dt, drivingForces)¶ Prepare state and model (temporarily) before solving a time-step
Synopsis:
[model, state] = model.prepareTimestep(state, state0, dt, drivingForces)
Description:
Prepare model and state just before the first call to
stepFunctionin a solution loop.Parameters: - model – Class instance
- state –
structrepresenting the current state of the solution variables to be updated. - problem –
LinearizedProblemADinstance that hasprimaryVariableswhich matchesdxin length and meaning. - dx – Cell-wise increments. These are typically output from
LinearSolverAD.solveLinearizedProblem. - forces – The forces used to produce the update. See
getDrivingForces.
Returns: - model – Updated model (non-persistent)
- state – Updated state with physically reasonable values.
Note
Any changes to the model are temporary for this specific step, as enforced by the NonLinearSolver. Any changes will not apply to the next time-step.
-
reduceState(model, state, removeContainers)¶ Reduce state to doubles, and optionally remove the property containers to reduce storage space
-
setProp(model, state, name, value)¶ Set named state property to given value
Synopsis:
state = model.setProp(state, 'PropertyName', value)
Parameters: - model – Class instance.
- state –
structholding the state of the nonlinear problem. - name – Name of the property to updated. See
getVariableField - value – The updated value that will be set.
Returns: state – Updated state
struct.Example:
% This will set state.pressure to 5 if the model knows of a % state field named pressure. If it is not known, it will % result in an error. state = struct('pressure', 0); state = model.setProp(state, 'pressure', 5);
-
solveAdjoint(model, solver, getState, getObj, schedule, gradient, stepNo)¶ Solve a single linear adjoint step to obtain the gradient
Synopsis:
gradient = model.solveAdjoint(solver, getState, ... getObjective, schedule, gradient, itNo)
Description:
This solves the linear adjoint equations. This is the backwards analogue of the forward mode
stepFunctioninPhysicalModelas well as thesolveTimestepmethod inNonLinearSolver.Parameters: - model – Class instance.
- solver – Linear solver to be used to solve the linearized system.
- getState –
Function handle. Should support the syntax:
state = getState(stepNo)
To obtain the converged state from the forward simulation for step
stepNo. - getObj –
Function handle providing the objective function for a specific step
stepNo:objfn = getObj(stepNo)
- schedule – Schedule used to compute the forward simulation.
- gradient – Current gradient to be updated. See outputs.
- stepNo – The current control step to be solved.
Returns: - gradient – The updated gradient.
- result – Solution of the adjoint problem.
- report – Report with information about the solution process.
See also
-
stepFunction(model, state, state0, dt, drivingForces, linsolver, nonlinsolver, iteration, varargin)¶ Perform a step that ideally brings the state closer to convergence
Synopsis:
[state, report] = model.stepFunction(state, state0, dt, ... forces, ls, nls, it)
Description:
Perform a single nonlinear step and report the progress towards convergence. The exact semantics of a nonlinear step varies from model to model, but the default behavior is to linearize the equations and solve a single step of the Newton-Rapshon algorithm for general nonlinear equations.
-
updateAfterConvergence(model, state0, state, dt, drivingForces)¶ Final update to the state after convergence has been achieved
Synopsis:
[state, report] = model.updateAfterConvergence(state0, state, dt, forces)
Description:
Update state based on nonlinear increment after timestep has converged. Defaults to doing nothing since not all models require this.
This function allows for the update of secondary variables instate after convergence has been achieved. This is especially useful for hysteretic parameters, where future function evaluations depend on the previous maximum value over all converged states.
Parameters: - model – Class instance.
- state –
structholding the current solution state. - forces – Driving forces used to execute the step. See
getDrivingForces.
Returns: - state – Updated
structholding the current solution state. - report – Report containing information about the update.
-
updateForChangedControls(model, state, forces)¶ Update model and state when controls/drivingForces has changed
Synopsis:
[model, state] = model.updateForChangedControls(state, forces)
Description:
Whenever controls change, this function should ensure that both model and state are up to date with the present set of driving forces.
Parameters: - model – Class instance.
- state –
structholding the current solution state. - forces – The new driving forces to be used in subsequent calls
to
getEquations. SeegetDrivingForces.
Returns: - model – Updated class instance.
- state – Updated
structholding the current solution state with accomodations made for any changed controls that provide e.g. primary variables.
-
updateState(model, state, problem, dx, forces)¶ Update the state based on increments of the primary values
Synopsis:
[state, report] = model.updateState(state, problem, dx, drivingForces)
Description:
Update the state’s primary variables (and any secondary quantities computing during the update process) based on a set of increments to each of the primary variables contained in
problem.primaryVariables.This function should ensure that values are within physically meaningful values and are meaningful so that the next call to
stepFunctioncan produce yet another set of reasonable increments in a process that eventually results in convergence.Parameters: - model – Class instance
- state –
structrepresenting the current state of the solution variables to be updated. - problem –
LinearizedProblemADinstance that hasprimaryVariableswhich matchesdxin length and meaning. - dx – Cell-wise increments. These are typically output from
LinearSolverAD.solveLinearizedProblem. - forces – The forces used to produce the update. See
getDrivingForces.
Returns: - state – Updated state with physically reasonable values.
- report – Struct with information about the update process.
Note
Specific properties can be manually updated with a variety of different functions. We trust the user and leave these functions as public. However, the main gateway to the update of state is through this function to ensure that all values are updated simultaneously. For many problems, updates can not be done separately and all changes in the primary variables must considered together for the optimal performance.
-
updateStateFromIncrement(model, state, dx, problem, name, relchangemax, abschangemax)¶ Update value in state, with optional limits on update magnitude
Synopsis:
state = model.updateStateFromIncrement(state, dx, problem, 'name') [state, val, val0] = model.updateStateFromIncrement(state, dx, problem, 'name', 1, 0.1)
Parameters: - model – Class instance.
- state – State `struct`to be updated.
- dx – Increments. Either a
cellarray matching the the primary variables ofproblem, or a single value. - problem –
LinearizedProblemused to obtain the increments. This input argument is only used ifdxis acellarray and can be replaced by a dummy value ifdxis a numerical type. - name – Name of the field to update, as supported by
model.getVariableField. - relchangemax – OPTIONAL. If provided, this will be interpreted as the maximum relative change in the variable allowed.
- abschangemax – OPTIONAL. If provided, this is the maximum absolute change in the variable allowed.
Returns: state – State with updated value.
Example:
% Update pressure with an increment of 10, without any limits, % resulting in the pressure being 110 after the update. state = struct('pressure', 10); state = model.updateStateFromIncrement(state, 100, problem, 'pressure') % Alternatively, we can use a relative limit on the update. In % the following, the pressure will be set to 11 as an immediate % update to 110 would violate the maximum relative change of % 0.1 (10 %) state = model.updateStateFromIncrement(state, 100, problem, 'pressure', .1)
Note
Relative limits such as these are important when working with tabulated and nonsmooth properties in a Newton-type loop, as the initial updates may be far outside the reasonable region of linearization for a complex problem. On the other hand, limiting the relative updates can delay convergence for smooth problems with analytic properties and will, in particular, prevent zero states from being updated, so use with care.
-
validateModel(model, varargin)¶ Validate model and check if it is ready for simulation
Synopsis:
model = model.validateModel(); model = model.validateModel(forces);
Description:
Validate that a model is suitable for simulation. If the missing or inconsistent parameters can be fixed automatically, an updated model will be returned. Otherwise, an error should occur.
Second input may be the forces struct argument. This function should NOT require forces arg to run, however.
Parameters: - model – Class instance to be validated.
- forces – (OPTIONAL): The forces to be used. Some models require
setup and configuration specific to the forces used.
This is especially important for the
FacilityModel, which implements the coupling between wells and the reservoir forReservoirModelsubclasses ofPhysicalModel.
Returns: model – Class instance. If returned, this model is ready for simulation. It may have been changed in the process.
-
validateState(model, state)¶ Validate state and check if it is ready for simulation
Synopsis:
state = model.validateState(state);
Description:
Validate the state for use with
model. Should check that required fields are present and of the right dimensions. If missing fields can be assigned default values, state is return with the required fields added. If reasonable default values cannot be assigned, a descriptive error should be thrown telling the user what is missing or wrong (and ideally how to fix it).Parameters: - model – Class instance for which
stateis intended as a valid state. - state –
structwhich is to be validated.
Returns: state –
struct. If returned, this state is ready for simulation withmodel. It may have been changed in the process.- model – Class instance for which
-
G= None¶ Grid. Can be empty.
-
nonlinearTolerance= None¶ Inf norm tolerance for checking residuals
-
operators= None¶ Operators used for construction of systems
-
stepFunctionIsLinear= None¶ Model step function is linear.
-
verbose= None¶ Verbosity from model routines
-
class
ReservoirModel(G, varargin)¶ Bases:
PhysicalModelBase class for physical models
Synopsis:
model = ReservoirModel(G, rock, fluid)
Description:
Extension of
PhysicalModelclass to accomodate reservoir-specific features such as fluid and rock as well as commonly used phases and variables.Parameters: - G – Simulation grid.
- rock – Valid rock used for the model.
- fluid – Fluid model used for the model.
Keyword Arguments: ‘property’ – Set property to the specified value.
Returns: Class instance.
See also
ThreePhaseBlackOilModel,TwoPhaseOilWaterModel,PhysicalModel-
addBoundaryConditionsAndSources(model, eqs, names, types, state, p, s, mob, rho, dissolved, components, forces)¶ Add in the boundary conditions and source terms to equations
Synopsis:
[eqs, state] = addBoundaryConditionsAndSources(model, eqs, names, types, state, ... p, sat, mob, rho, ... rs, components, ... drivingForces);
Parameters: - model – Class instance.
- eqs – Cell array of equations that are to be updated.
- names – The names of the equations to be updated. If phase-pseudocomponents are to be used, the names must correspond to some combination of “water”, “oil”, “gas” if no special component treatment is to be introduced.
- types – Cell array with the types of “eqs”. Note that these types must be ‘cell’ where source terms is to be added.
- src – Struct containing all the different source terms that were computed and added to the equations.
- p – Cell array of phase pressures.
- s – Cell array of phase saturations.
- mob – Cell array of phase mobilities
- rho – Cell array of phase densities
- dissolved – Cell array of dissolved components for black-oil style pseudocompositional models.
- components – Cell array of equal length to the number of
components. The exact representation may vary
based on the model, but the respective
sub-component is passed onto
addComponentContributions. - forces – DrivingForces struct (see
getValidDrivingForces) containing (possibily empty)srcandbcfields.
Returns: - eqs – Equations with corresponding source terms added.
- state – Reservoir state. Can be modified to store e.g. boundary fluxes due to boundary conditions.
- src – Normalized struct containing the source terms used.
Note
This function accomodates both the option of black-oil pseudocomponents (if the model equations are named “oil”, “gas” or water) and true components existing in multiple phases. Mixing the two behaviors can lead to unexpected source terms.
-
addComponentContributions(model, cname, eq, component, src, force)¶ For a given component conservation equation, compute and add in source terms for a specific source/bc where the fluxes have already been computed.
Parameters: - model – (Base class, automatic)
- cname – Name of the component. Must be a property known to the
model itself through
getPropandgetVariableField. - eq – Equation where the source terms are to be added. Should be one value per cell in the simulation grid (model.G) so that the src.sourceCells is meaningful.
- component – Cell-wise values of the component in question. Used for outflow source terms only.
- src – Source struct containing fields for fluxes etc. Should
be constructed from force and the current reservoir
state by
computeSourcesAndBoundaryConditionsAD. - force – Force struct used to produce src. Should contain the field defining the component in question, so that the inflow of the component through the boundary condition or source terms can accurately by estimated.
-
static
adjustStepFromSatBounds(s, ds)¶ Ensure that cellwise increment for each phase is done with the same length, in a manner that avoids saturation violations.
-
compVarIndex(model, name)¶ Find the index of a component variable by name
Synopsis:
index = model.compVarIndex('co2')
Parameters: - model – Class instance
- name – Name of component.
Returns: index – Index of the component for this model. Empty if saturation was not known to model.
-
evaluateRelPerm(model, sat, varargin)¶ Evaluate relative permeability corresponding to active phases
Synopsis:
% Single-phase water model krW = model.evaluateRelPerm({sW}); % Two-phase oil-water model [krW, krO] = model.evaluateRelPerm({sW, sO}); % Two-phase water-gas model [krW, krG] = model.evaluateRelPerm({sW, sG}); % Three-phase oil-water-gas model [krW, krO, krG] = model.evaluateRelPerm({sW, sO, sG});
Parameters: - model – The model
- sat – Cell array containing the saturations for all active phases.
Returns: varargout – One output argument per phase present, corresponding to evaluated relative permeability functions for each phase in the canonical ordering.
See also
relPermWOG,relPermWO,relPermOG,relPermWG
-
getActivePhases(model)¶ Get active flag for MRST’s canonical phase ordering (WOG)
Synopsis:
[act, indices] = model.getActivePhases();
Parameters: model – Class instance
Returns: - isActive – Total number of known phases array with booleans indicating if that phase is present. MRST uses a ordering of water, oil and then gas.
- phInd – Indices of the phases present. For instance, if
water and gas are the only ones present,
phInd = [1, 3]
-
getComponentNames(model)¶ #ok Get the names of components for the model .. rubric:: Synopsis:
names = model.getComponentNames();
Parameters: model – Class instance. Returns: names – Cell array of component names in no particular order.
-
getConvergenceValues(model, problem, varargin)¶ Check convergence criterion. Relies on
FacilityModelto check wells.
-
getExtraWellContributions(model, well, wellSol0, wellSol, q_s, bh, packed, qMass, qVol, dt, iteration)¶ This function is called by the well model (base class: SimpleWell) during the assembly of well equations and addition of well source terms. The purpose of this function, given the internal variables of the well model, is to compute the additional closure equations and source terms that the model requires. For instance, if the model contains different components that require special treatment (see for example the implementation of this function in
ad_eor.models.OilWaterPolymerModelin thead-eormodule), this function should assemble any additional equations and corresponding source terms. It is also possible to add source terms without actually adding well equations.Returns: - compEqs – A cell array of additional equations added to the system to account for the treatment of components etc in the well system.
- compSrc – Cell array of component source terms, ordered and with
the same length as the output from
ReservoirModel.getComponentNames. - eqNames – Names of the added equations. Must correspond to the
same entries as
getExtraWellEquationNames(but does not have to maintain the same ordering).
Note
Input arguments are intentionally undocumented and subject to change. Please see
SimpleWellfor details.
-
getExtraWellEquationNames(model)¶ Get the names and types of extra well equations in model
Synopsis:
[names, types] = model.getExtraWellEquationNames();
Parameters: model – Base class.
Returns: - names – Cell array of additional well equations.
- types – Cell array of corresponding types to
names.
See also
getExtraWellContributions
-
getExtraWellPrimaryVariableNames(model)¶ Get the names of extra well primary variables required by model.
Synopsis:
names = model.getExtraWellPrimaryVariableNames();
Parameters: model – Class instance Returns: names – Cell array of named primary variables. See also
getExtraWellContributions
-
getGravityGradient(model)¶ Get the discretized gravity contribution on faces
Synopsis:
gdz = model.getGravityGradient();
Parameters: model – Class instance Returns: g – One entry of the gravity contribution per face in the grid. Does not necessarily assume that gravity is aligned with one specific direction. See also
-
getGravityVector(model)¶ Get the gravity vector used to instantiate the model
Synopsis:
g = model.getGravityVector();
Parameters: model – Class instance Returns: g – model.G.griddimlong vector representing the gravity accleration constant along increasing depth.See also
-
getMaximumTimestep(model, state, state0, dt0, drivingForces)¶ Define the maximum allowable time-step based on physics or discretization choice
-
getPhaseIndex(model, phasename)¶ Query the index of a phase in the model’s ordering
Synopsis:
index = model.getPhaseNames();
Parameters: - model – Class instance
- phasename – The name of the phase to be found.
Returns: index – Index of phase
phasename
-
getPhaseIndices(model)¶ Get the active phases in canonical ordering
-
getPhaseNames(model)¶ Get short and long names of the present phases.
Synopsis:
[phNames, longNames] = model.getPhaseNames();
Parameters: model – Class instance
Returns: - phNames – Cell array containing the short hanes (‘W’, ‘O’, G’) of the phases present
- longNames – Longer names (‘water’, ‘oil’, ‘gas’) of the phases present.
-
getScalingFactorsCPR(model, problem, names, solver)¶ #ok Get scaling factors for CPR reduction in
CPRSolverADParameters: - model – Class instance
- problem –
LinearizedProblemADwhich is intended for CPR preconditioning. - names – The names of the equations for which the factors are to be obtained.
- solver – The
LinearSolverADclass requesting the scaling factors.
Returns: scaling – Cell array with either a scalar scaling factor for each equation, or a vector of equal length to that equation.
- SEE ALSO
CPRSolverAD
-
getSurfaceDensities(model)¶ Get the surface densities of the active phases in canonical ordering (WOG, with any inactive phases removed).
Returns: rhoS – pvt x n double array of surface densities.
-
getValidDrivingForces(model)¶ Get valid forces. This class adds support for wells, bc and src.
-
getVariableField(model, name, varargin)¶ Map variables to state field.
-
insertWellEquations(model, eqs, names, types, wellSol0, wellSol, wellVars, wellMap, p, mob, rho, dissolved, components, dt, opt)¶ Add in the effect of wells to a system of equations, by adding corresponding source terms and augmenting the system with additional equations for the wells.
Parameters: - eqs – Cell array of equations that are to be updated.
- names – The names of the equations to be updated. If phase-pseudocomponents are to be used, the names must correspond to some combination of “water”, “oil”, “gas” if no special component treatment is to be introduced.
- types – Cell array with the types of “eqs”. Note that these types must be ‘cell’ where source terms is to be added.
- src – Struct containing all the different source terms that were computed and added to the equations.
- various – Additional input arguments correspond to standard reservoir-well coupling found in `FacilityModel.
-
static
relPermOG(so, sg, f, varargin)¶ Two-phase oil-gas relative permeability function
Synopsis:
[krO, krG] = model.relPermOG(so, sg, f);
Parameters: - sw – Water saturation
- sg – Gas saturation
- f –
Struct representing the field. Fields that are used:
krO: Oil relperm function of oil saturation.krOG: Oil-gas relperm function of gas saturation. This function is only used ifkrOis not found.krG: Gas relperm function of gas saturation.
Returns: - krO – Oil relative permeability.
- krG – Gas relative permeability
Note
This function should typically not be called directly as its interface is subject to change. Instead, use
evaluateRelPerm.
-
static
relPermWG(sw, sg, f, varargin)¶ Two-phase water-gas relative permeability function
Synopsis:
[krW, krG] = model.relPermWG(sw, sg, f);
Parameters: - sw – Water saturation
- sg – Gas saturation
- f –
Struct representing the field. Fields that are used:
krW: Water relperm function of water saturation.krG: Gas relperm function of gas saturation.
Returns: - krW – Water relative permeability.
- krG – Gas relative permeability
Note
This function should typically not be called directly as its interface is subject to change. Instead, use
evaluateRelPerm.
-
static
relPermWO(sw, so, f, varargin)¶ Two-phase water-oil relative permeability function
Synopsis:
[krW, krO] = model.relPermWO(sw, so, f);
Parameters: - sw – Water saturation
- so – Oil saturation
- f –
Struct representing the field. Fields that are used:
krW: Water relperm function of water saturation.krO: Oil relperm function of oil saturation.krOW: Oil-water relperm function of oil saturation. Only used ifkrOis not found.
Returns: - krW – Water relative permeability.
- krO – Oil relative permeability.
Note
This function should typically not be called directly as its interface is subject to change. Instead, use
evaluateRelPerm.
-
static
relPermWOG(sw, so, sg, f, varargin)¶ Three-phase water-oil-gas relative permeability function
Synopsis:
[krW, krO, krG] = model.relPermWOG(sw, so, sg, f);
Parameters: - sw – Water saturation
- so – Oil saturation
- sg – Gas saturation
- f –
Struct representing the field. Fields that are used:
krW: Water relperm function of water saturation.krOW: Oil-water relperm function of oil saturation.krOG: Oil-gas relperm function of oil saturation.krG: Gas relperm function of gas saturation.sWcon: Connate water saturation. OPTIONAL.
Returns: - krW – Water relative permeability.
- krO – Oil relative permeability.
- krG – Gas relative permeability
Note
This function should typically not be called directly as its interface is subject to change. Instead, use
evaluateRelPerm.
-
satVarIndex(model, name)¶ Find the index of a saturation variable by name
Synopsis:
index = model.satVarIndex('water')
Parameters: - model – Class instance
- name – Name of phase.
Returns: index – Index of the phase for this model. Empty if saturation was not found.
-
setPhaseData(model, state, data, fld, subs)¶ Store phase data in state for further output.
Synopsis:
state = model.setPhaseData(state, data, 'someField') state = model.setPhaseData(state, data, 'someField', indices)
Description:
Utility function for storing phase data in the state. This is used for densities, fluxes, mobilities and so on when requested from the simulator.
Parameters: - model – Class instance
- data – Cell array of data to be stored. One entry per active
phase in
model. - fld – The field to be stored.
- subs –
OPTIONAL. The subset for which phase data is to be stored. Must be a valid index of the type
data.(fld)(subs, someIndex) = data{i}
for all i. Defaults to all indices.
Returns: state – state with updated
fld.
-
setupOperators(model, G, rock, varargin)¶ Set up default discretization operators and other static props
Synopsis:
model = model.setupOperators(G, rock)
Description:
This function calls the default set of discrete operators as implemented by
setupOperatorsTPFA. The default operators use standard choices for reservoir simulation similar to what is found in commercial simulators: Single-point potential upwind and two-point flux approximation.Parameters: - model – Class instance.
- G – The grid used for computing the operators. Must have
geometry information added (typically from
computeGeometry). Although this is a property on the model, we allow for passing of a different grid for the operator setup since this is useful in some workflows. - rock – Rock structure. See
makeRock.
Returns: model – Model with updated
operatorsproperty.Note
This function is called automatically during class construction.
-
splitPrimaryVariables(model, vars)¶ Split cell array of primary variables into grouping .. rubric:: Synopsis:
[restVars, satVars, wellVars] = model.splitPrimaryVariables(vars)
Description:
Split a set of primary variables into three groups: Well variables, saturation variables and the rest. This is useful because the saturation variables usually are updated together, and the well variables are a special case.
Parameters: - model – Class instance.
- vars – Cell array with names of primary variables
Returns: - restVars – Names of variables that are not saturations or belong to the wells.
- satVars – Names of the saturation variables present in
vars. - wellVars – Names of the well variables present in
vars
-
storeBoundaryFluxes(model, state, qW, qO, qG, forces)¶ Store integrated phase fluxes on boundary in state.
Synopsis:
Three-phase case state = model.storeBoundaryFluxes(state, vW, vO, vG, forces); % Only water and gas in model: state = model.storeBoundaryFluxes(state, vW, [], vG, forces);
Parameters: - model – Class instance
- vW – Water fluxes, one value per BC interface.
- vO – Oil fluxes, one value per BC interface.
- vG – Gas fluxes, one value per BC interface.
- forces –
drivingForcesstruct containing any boundary conditions used to obtain fluxes.
Returns: state – State with stored values
See also
storeFluxes
-
storeDensity(model, state, rhoW, rhoO, rhoG)¶ Store phase densities per-cell in state.
Synopsis:
Three-phase case state = model.storeDensity(state, rhoW, rhoO, rhoG); % Only water and gas in model: state = model.storeDensity(state, rhoW, [], rhoG);
Parameters: - model – Class instance
- rhoW – Water densities, one value per internal interface.
- rhoO – Oil densities, one value per internal interface.
- rhoG – Gas densities, one value per internal interface.
Returns: state – State with stored values
See also
storeMobilities
-
storeFluxes(model, state, vW, vO, vG)¶ Store integrated internal phase fluxes in state.
Synopsis:
Three-phase case state = model.storeFluxes(state, vW, vO, vG); % Only water and gas in model: state = model.storeFluxes(state, vW, [], vG);
Parameters: - model – Class instance
- vW – Water fluxes, one value per internal interface.
- vO – Oil fluxes, one value per internal interface.
- vG – Gas fluxes, one value per internal interface.
Returns: state – State with stored values
See also
storeBoundaryFluxes
-
storeMobilities(model, state, mobW, mobO, mobG)¶ Store phase mobility per-cell in state.
Synopsis:
Three-phase case state = model.storebfactors(state, mobW, mobO, mobG); % Only water and gas in model: state = model.storebfactors(state, mobW, [], mobG);
Parameters: - model – Class instance
- mobW – Water mobilities, one value per internal interface.
- mobO – Oil mobilities, one value per internal interface.
- mobG – Gas mobilities, one value per internal interface.
Returns: state – State with stored values
See also
storeMobilities
-
storeUpstreamIndices(model, state, upcw, upco, upcg)¶ Store upstream indices for each phase in state.
Synopsis:
Three-phase case state = model.storeUpstreamIndices(state, upcw, upco, upcg); % Only water and gas in model: state = model.storeUpstreamIndices(state, upcw, [], upcg);
Parameters: - model – Class instance
- upcw – Water upwind indicator, one value per internal interface.
- upco – Oil upwind indicator, one value per internal interface.
- upcg – Gas upwind indicator, one value per internal interface.
Returns: state – State with stored values
See also
storeBoundaryFluxes,storeFluxes
-
storebfactors(model, state, bW, bO, bG)¶ Store phase reciprocal FVF per-cell in state.
Synopsis:
Three-phase case state = model.storebfactors(state, bW, bO, bG); % Only water and gas in model: state = model.storebfactors(state, bW, [], bG);
Parameters: - model – Class instance
- bW – Water reciprocal FVF, one value per internal interface.
- bO – Oil reciprocal FVF, one value per internal interface.
- bG – Gas reciprocal FVF, one value per internal interface.
Returns: state – State with stored values
See also
storeDensities
-
updateAfterConvergence(model, state0, state, dt, drivingForces)¶ Generic update function for reservoir models containing wells.
-
updateForChangedControls(model, state, forces)¶ Called whenever controls change.
Note
The addition this class makes is also updating the well solution and the underlying
FacilityModelclass instance.
-
updateSaturations(model, state, dx, problem, satVars)¶ Update of phase-saturations
Synopsis:
state = model.updateSaturations(state, dx, problem, satVars)
Description:
Update saturations (likely state.s) under the constraint that the sum of volume fractions is always equal to 1. This assumes that we have solved for n - 1 phases when n phases are present.
Parameters: - model – Class instance
- state – State to be updated
- dx – Cell array of increments, some of which correspond to saturations
- problem –
LinearizedProblemADclass instance from whichdxwas obtained. - satVars – Cell array with the names of the saturation variables.
Returns: state – Updated state with saturations within physical constraints.
See also
splitPrimaryVariables
-
updateState(model, state, problem, dx, drivingForces)¶ Generic update function for reservoir models containing wells.
-
validateModel(model, varargin)¶ Validate model.
-
validateState(model, state)¶ Validate initial state.
-
FacilityModel= None¶ Facility model used to represent wells
-
FlowPropertyFunctions= None¶ Grouping for flow properties
-
FluxDiscretization= None¶ Grouping for flux discretization
-
dpMaxAbs= None¶ Maximum absolute pressure change
-
dpMaxRel= None¶ Maximum relative pressure change
-
dsMaxAbs= None¶ Maximum absolute saturation change
-
extraStateOutput= None¶ Write extra data to states. Depends on submodel type.
-
extraWellSolOutput= None¶ Output extra data to wellSols: GOR, WOR, …
-
fluid= None¶ The fluid model. See
initSimpleADIFluid,initDeckADIFluid
-
gas= None¶ Indicator showing if the vapor/gas phase is present
-
gravity= None¶ Vector for the gravitational force
-
inputdata= None¶ Input data used to instantiate the model
-
maximumPressure= None¶ Maximum pressure allowed in reservoir
-
minimumPressure= None¶ Minimum pressure allowed in reservoir
-
oil= None¶ Indicator showing if the liquid/oil phase is present
-
outputFluxes= None¶ Store integrated fluxes in state.
-
useCNVConvergence= None¶ Use volume-scaled tolerance scheme
-
water= None¶ Indicator showing if the aqueous/water phase is present
-
class
WrapperModel(parentModel, varargin)¶ Bases:
PhysicalModelWrapper model which can be inherited for operations on the parent model.
-
prepareReportstep(model, state, state0, dt, drivingForces)¶ Prepare state and model (temporarily) before solving a report-step
-
prepareTimestep(model, state, state0, dt, drivingForces)¶ Prepare state and model (temporarily) before solving a time-step
-
Facility and wells¶
-
Contents¶ FACILITIES
- Files
- combineMSwithRegularWells - Combine regular and MS wells, accounting for missing fields computeWellContributionsSingleWell - Main internal function for computing well equations and source terms ExtendedFacilityModel - FacilityModel - A model coupling facilities and wells to the reservoir MultisegmentWell - Derived class implementing multisegment wells nozzleValve - Nozzle valve model packPerforationProperties - Extract variables corresponding to a specific well selectFacilityFromDeck - Pick FacilityModel from input deck setupMSWellEquationSingleWell - Setup well residual equations for multi-segmented wells - and only those. setupWellControlEquationsSingleWell - Setup well controll (residual) equations SimpleWell - Base class implementing a single, instantaneous equilibrium well model unpackPerforationProperties - Unpack the properties extracted by packPerforationProperties. Internal function. wellBoreFriction - Empricial model for well-bore friction UniformFacilityModel - Simplified facility model which is sometimes faster reorderWellPerforationsByDepth - Undocumented Utility Function
-
class
FacilityModel(reservoirModel, varargin)¶ Bases:
PhysicalModelA model coupling facilities and wells to the reservoir
Synopsis:
model = FacilityModel(reservoirModel)
Description:
The
FacilityModelis the layer between aReservoirModeland the facilities. The facilities consist of a number of different wells that are implemented in their own subclasses.Different wells have different governing equations, primary variables and convergence criterions. This class seamlessly handles wells appearing and disappearing, controls changing and even well type changing.
Parameters: resModel – ReservoirModelderived class the facilities are coupled to.Keyword Arguments: ‘property’ – Set property to the specified value. Returns: model – Class instance of FacilityModel.See also
ReservoirModel,PhysicalModel,SimpleWell-
checkFacilityConvergence(model, problem)¶ Check if facility and submodels has converged
Synopsis:
[convergence, values, names, evaluated] = model. checkFacilityConvergence(problem)
Parameters: - model – Class instance.
- problem –
LinearizedProblemADto be checked for convergence.
Returns: - conv – Array of convergence flags for all tests.
- vals – Values checked to obtain
conv. - names – The names of the convergence checks/equations.
- eval – Logical array into problem.equations indicating which residual equations we have actually checked convergence for.
-
getActiveWellCells(model, wellSol)¶ Get the perforated cells in active wells
Synopsis:
wc = model.getActiveWellCells()
Parameters: model – Facility model class instance. Returns: wc – Array of well cells that are active, and belong to active wells.
-
getAllPrimaryVariables(model, wellSol)¶ Get all primary variables (basic + extra)
Synopsis:
[variables, names, wellmap] = model.getAllPrimaryVariables(wellSol)
Description:
Gets all primary variables, both basic (rates, bhp) and added variables (added by different wells and from the model itself).
Parameters: - model –
FacilityModelclass instance - wellSol – The wellSol struct
Returns: - names – Cell array. Each cell is a string with the name of an variable.
- variables – Column of cells. Each element, variables{i}, is a vector given the value corresponding to variable with name names{i}. This vector is composed of stacked values over all the wells that contains this variable.
- wellmap – A combined struct containing mappings for both the standard and extra primary variables.
See also
getBasicPrimaryVariables,getExtraPrimaryVariables- model –
-
getBasicPrimaryVariableNames(model)¶ Get the names of the basic primary variables present in all wells
Synopsis:
names = model.getBasicPrimaryVariableNames();
Description:
Get a list of the basic names of primary variables that will be required to solve a problem with the current set of wells. The basic primary variables are always present in MRST, and correspond to the phase rates for each phase present, as well as the bottom-hole pressures. This ensures that all solvers have a minimum feature set for well controls.
Parameters: model – FacilityModelclass instanceReturns: names – Cell array of the names of the basic primary variables.
-
getBasicPrimaryVariables(model, wellSol)¶ Get the basic primary variables common to all well models.
Synopsis:
[vars, names, map] = model.getBasicPrimaryVariables(wellSol)
Description:
Get phase rates for the active phases and the bhp. In addition, the map contains indicators used to find the phase rates and BHP values in “variables” since these are of special importance to many applications and are considered canonical (i.e. they are always solution variables in MRST, and functions can assume that they will always be found in the variable set for wells).
Parameters: model –
FacilityModelclass instanceReturns: - variables – Cell array of the primary variables.
- names – Cell array with the names of the primary variables.
- map – Struct with details on which variables correspond to ordered phase rates and the bottom hole pressures.
-
getEquations(model, state0, state, dt, drivingForces, varargin)¶ Get stand-alone equations for the wells
Synopsis:
[problem, state] = model.getEquations(state0, state, dt, drivingForces, varargin)
Description:
The well equations can be solved as a separate nonlinear system with the reservoir as a fixed quantity. This is useful for debugging.
Parameters: - model –
FacilityModelinstance. - wellSol0 – wellSol struct at previous time-step.
- wellSol – wellSol struct at current time-step.
- dt – Time-step.
- forces – Forces struct for the wells.
Keyword Arguments: - ‘resOnly’ – If supported by the equations, this flag will result in only the values of the equations being computed, omitting any Jacobians.
- ‘iteration’ – The nonlinear iteration number. This can be provided so that the underlying equations can account for the progress of the nonlinear solution process in a limited degree, for example by updating some quantities only at the first iteration.
Returns: - problem – Instance of the wrapper class
LinearizedProblemADcontaining the residual equations as well as other useful information. - state – The equations are allowed to modify the system state, allowing a limited caching of expensive calculations only performed when necessary.
- model –
-
getExtraPrimaryVariables(model, wellSol)¶ Get additional primary variables (not in the basic set)
Synopsis:
[variables, names, wellmap] = model.getExtraPrimaryVariables(wellSol)
Description:
Get extra primary variables are variables required by more advanced wells that are in addition to the basic facility variables (rates + bhp).
Parameters: - model –
FacilityModelclass instance - wellSol – The wellSol struct
Returns: - names – Column of cells. Each cell is a string with the name of an extra-variable.
- variables – Column of cells. Each element, variables{i}, is a vector given the value corresponding to extra-variable with name names{i}. This vector is composed of stacked values over all the wells that contains this extra-variable.
- wellmap – The facility model contains the extra-variables of all
the well models that are used. Let us consider the well
with well number wno (in the set of active wells), then
the Well model is belongs to has its own
extra-variables (a subset of those of the Facility
model). We consider the j-th extra-variable of
the Well model. Then,
i = extraMap(wno, j)says that this extra-variable corresponds tonames{i}.
See also
getBasicPrimaryVariables- model –
-
getIndicesOfActiveWells(model, wellSol)¶ Get indices of active wells
Synopsis:
actIx = model.getIndicesOfActiveWells(wellSol);
Parameters: - model –
FacilityModelclass instance - wellSol – The wellSol struct
Returns: actIx – The indices of the active wells in the global list of all wells (active & inactive)
- model –
-
getNumberOfActiveWells(model, wellSol)¶ Get number of wells active initialized in facility
Synopsis:
W = model.getNumberOfActiveWells();
Parameters: model – FacilityModelclass instanceReturns: nwell – Number of active wells
-
getNumberOfWells(model)¶ Get number of wells initialized in facility
Synopsis:
W = model.getNumberOfWells();
Parameters: model – FacilityModelclass instanceReturns: nwell – Number of wells
-
getPrimaryVariableNames(model)¶ Get the names of primary variables present in all wells
Synopsis:
names = model.getPrimaryVariableNames();
Description:
Get a list of the names of primary variables that will be required to solve a problem with the current set of wells.
Parameters: model – FacilityModelclass instanceReturns: names – Cell array of the names of the primary variables.
-
getValidDrivingForces(model)¶ Get valid driving forces.
-
getWellCells(model, subs)¶ Get the perforated cells of all wells, regardless of status
Synopsis:
wc = model.getWellCells()
Parameters: model – Facility model class instance. Returns: wc – Array of well cells
-
getWellContributions(model, wellSol0, wellSol, wellvars, wellMap, p, mob, rho, dissolved, comp, dt, iteration)¶ Get sources, well equations and updated wellSol
Synopsis:
[sources, wellSystem, wellSol] = fm.getWellContributions(... wellSol0, wellSol, wellvars, wellMap, p, mob, rho, dissolved, comp, dt, iteration)
Description:
Get the source terms due to the wells, control and well equations and updated well sol. Main gateway for adding wells to a set of equations.
Parameters: - model – Facility model class instance.
- wellSol0 – wellSol struct at previous time-step.
- wellSol – wellSol struct at current time-step.
- wellvars – Well variables. Output from
getAllPrimaryVariables. - wellMap – Well mapping. Output from
getAllPrimaryVariables. - p – Pressure defined in all cells of the underlying
ReservoirModel. Normally, this is the oil pressure. - mob – Cell array of phase mobilities defined in all cells of the reservoir.
- rho – Cell array of phase densities defined in all cells of the reservoir.
- dissolved – Black-oil style dissolution. See
ad_blackoil.models.ThreePhaseBlackoilModel.getDissolutionMatrix. - comp – Cell array of components in the reservoir.
- dt – The time-step.
- iteration – The current nonlinear iteration for which the sources are to be computed.
Returns: - sources – Struct containing source terms for phases, components and the corresponding cells.
- wellSystem – Struct containing the well equations (reservoir to wellbore, and control-equations).
- wellSol – Updated wellSol struct.
-
getWellStatusMask(model, wellSol)¶ Get status mask for active wells
Synopsis:
act = model.getWellStatusMask(wellSol);
Description:
Get the well status of all wells. The status is true if the well is present and active. Wells can be disabled in two ways: Their status flag can be set to false in the well struct, or the wellSol.status flag can be set to false by the simulator itself.
Parameters: - model –
FacilityModelclass instance - wellSol – The wellSol struct
Returns: act – Array with equal length to the total number of wells, with booleans indicating if a specific well is currently active.
- model –
-
getWellStruct(model, subs)¶ Get the well struct representing the current set of wells
Synopsis:
W = model.getWellStruct();
Parameters: model – FacilityModelclass instanceReturns: W – Standard well struct.
-
getWellVariableMap(model, wf, ws)¶ Get mapping indicating which variable belong to each well
Synopsis:
isVarWell = model.getWellVariableMap('someVar', wellSol)
Parameters: - model – Class instance of
FacilityModel - wf – String of variable for which the mapping will be generated.
- ws – Current wellSol.
Returns: isVarWell – Array equal in length to the total number of variables with name
wf. The entries correspond to which well owns that specific variable number. This allows multiple wells to have for example bottom-hole pressures as a variable, without having to split them up by name in the reservoir equations.- model – Class instance of
-
static
handleRepeatedPerforatedcells(wc, varargin)¶ Handle multiple wells perforated in the same cells
Synopsis:
[wc, src1, src2] = handleRepeatedPerforatedcells(wc, src1, src2);
Description:
This function treats repeated indices in wc (typically due to multiple wells intersecting a single cell). The output will have no repeats in wc, and add together any terms in cqs.
Parameters: - wc – Well cells with possible repeats.
- varargin – Any number of arrays that should be processed to account for repeated entries.
Returns: - wc – Well cells with repeats removed.
- varargout – Variable inputs processed to fix repeated indices.
-
setWellSolStatistics(model, ws, sources)¶ Add statistics to wellSol (wcut, gor, …)
Synopsis:
wellSol = model.setWellSolStatistics(wellSol, sources)
Parameters: - model – Facility model class instance.
- wellSol – wellSol struct at current time-step.
- sources – Source struct from
getWellContributions.
Returns: wellSol – Updated wellSol struct where additional useful information has been added
-
setupWells(model, W, wellmodels)¶ Set up well models for changed controls or the first simulation step.
Parameters: W – Well struct (obtained from e.g. addWellorprocessWells)Keyword Arguments: wellmodels – Cell array of equal length to W, containing class instances for each well (e.g. SimpleWell,MultisegmentWell, or classes derived from these). If not provided, well models be constructed from the input.Returns: model – Updated FacilityModelinstance ready for use with wells of typeW.
-
updateAfterConvergence(model, state0, state, dt, drivingForces)¶ Generic update function for reservoir models containing wells.
-
updateState(model, state, problem, dx, drivingForces)¶ Update state.
-
updateWellSol(model, wellSol, problem, dx, drivingForces, restVars)¶ Update the wellSol based on increments
Synopsis:
[wellSol, restVars] = model.updateWellSol(wellSol, problem, dx, forces, restVars)
Parameters: - model – Facility model class instance
- wellSol – Well solution struct
- problem – Linearized problem used to produce dx.
- dx – Increments corresponding to
problem.primaryVariables - forces – Boundary condition struct
- restVars – Variables that have not yet been updated.
Returns: - state – Updated Well solution struct
- restVars – Variables that have not yet been updated.
-
updateWellSolAfterStep(model, wellSol, wellSol0)¶ Update wellSol after step (check for closed wells, etc)
Synopsis:
wellSol = model.updateWellSolAfterStep(wellSol, wellSol0)
Parameters: - model – Facility model class instance.
- wellSol0 – wellSol struct at previous time-step.
- wellSol – wellSol struct at current time-step.
Returns: wellSol – Updated wellSol struct.
-
validateState(model, state)¶ Validate state.
-
FacilityFluxDiscretization= None¶ Convergence tolerance for BHP-type controls
-
ReservoirModel= None¶ The model instance the FacilityModel is coupled to
-
VFPTablesInjector= None¶ Injector VFP Tables. EXPERIMENTAL.
-
VFPTablesProducer= None¶ Producer VFP Tables. EXPERIMENTAL.
-
WellModels= None¶ Cell array of instantiated wells
-
addedEquationNames= u'{}'¶ Canonical list of additional equations
-
addedEquationTypes= u'{}'¶ Canonical list of the types of the added equations
-
addedPrimaryVarNames= u'{}'¶ Canonical list of all extra primary variables added by the wells
-
addedPrimaryVarNamesIsFromResModel= u'[]'¶ Indicator, per primary variable, if it was added by the reservoir model (true) or if it is from the well itself (false)
-
toleranceWellMS= None¶ Convergence tolerance for multisegment wells
-
toleranceWellRate= None¶ Convergence tolerance for rate-type controls
-
-
class
MultisegmentWell(W, varargin)¶ Bases:
SimpleWellDerived class implementing multisegment wells
Synopsis:
wm = MultisegmentWell(W)
Description:
This well extends SimpleWell to general multisegment wells. These wells can take on complex topological structures, including loops for e.g. annular flow modelling.
Parameters: W – Well struct. See addWellandprocessWells. Should have been converted into a multisegment well usingconvert2MSWell.Keyword Arguments: ‘property’ – Set property to the specified value. Returns: model – Class instance of MultisegmentWell.See also
convert2MSWell,SimpleWell,-
computeWellEquations(well, wellSol0, wellSol, resmodel, q_s, bh, packed, dt, iteration)¶ Node pressures for the well
-
ensureWellSolConsistency(well, ws)¶ #ok guarantees that the sum of rW, rO, rG remains equal to one.
-
getVariableField(model, name, varargin)¶ Get the index/name mapping for the model (such as where pressure or water saturation is located in state)
-
-
class
SimpleWell(W, varargin)¶ Bases:
PhysicalModelBase class implementing a single, instantaneous equilibrium well model
Synopsis:
wm = SimpleWell(W)
Description:
Base class for wells in the AD-OO framework. The base class is also the default well implementation. For this will model, the assumptions are that the well-bore flow is rapid compared to the time-steps taken by the reservoir simulator, making instantaneous equilibrium and mixing in the well-bore a reasonable assumption.
Parameters: W – Well struct. See addWellandprocessWells.Keyword Arguments: ‘property’ – Set property to the specified value. Returns: model – Class instance of SimpleWell.See also
FacilityModel,MultisegmentWell,-
SimpleWell(W, varargin)¶ Class constructor
-
computeComponentContributions(well, wellSol0, wellSol, resmodel, q_s, bh, packed, qMass, qVol, dt, iteration)¶ Compute component equations and component source terms
See also
ad_core.models.ReservoirModel.getExtraWellContributions
-
computeWellEquations(well, wellSol0, wellSol, resmodel, q_s, bh, packed, dt, iteration)¶ Compute well equations and well phase/pseudocomponent source terms
-
ensureWellSolConsistency(well, ws)¶ #ok Run after the update step to ensure consistency of wellSol
-
getExtraEquationNames(well, resmodel)¶ Returns the names and types of the additional equation names this well model introduces.
Synopsis:
[names, types] = well.getExtraEquationNames(model)
Description:
We have two options: Either the well itself can add additional equations (modelling e.g. flow in the well-bore) or the reservoir can add additional equations (typically for additional components)
Parameters: - well – Well class instance
- resmodel –
ReservoirModelclass instance.
Returns: - names – Names of additional equations.
- types – Type hints for the additional equations.
-
getExtraPrimaryVariableNames(well, resmodel)¶ Get additional primary variables added by this well.
Synopsis:
[names, fromRes] = well.getExtraPrimaryVariableNames(model)
Description:
Additional primary variables in this context are variables that are not the default MRST values (surface rates for each pseudocomponent/phase and bottomhole pressure).
In addition, this function returns a indicator per variable if it was added by the reservoir model, or the well model.
Parameters: - well – Well class instance
- resmodel –
ReservoirModelclass instance.
Returns: - names – Names of additional primary variables.
- fromRes – Boolean array indicating if the added variables originate from the well, or the reservoir.
-
getExtraPrimaryVariables(well, wellSol, resmodel)¶ Returns the values and names of extra primary variables added by this well.
Synopsis:
[names, types] = well.getExtraEquationNames(model)
Parameters: - well – Well class instance
- resmodel –
ReservoirModelclass instance.
Returns: - vars – Cell array of extra primary variables
- names – Cell array with names of extra primary variables
See also
getExtraPrimaryVariableNames
-
getVariableCounts(wm, fld)¶ Get number of primary variables of a specific type for well
Synopsis:
counts = wellmodel.getVariableCounts('bhp')
Description:
Should return the number of primary variables added by this well for field “fld”. The simple base class only uses a single variable to represent any kind of well field, but in e.g.
MultisegmentWell, this function may return values larger than 1.Note
A value of zero should be returned for a unknown field.
Parameters: - wellmodel – Well model class instance.
- fld – Primary variable name.
Returns: counts – Number of variables of this type required by the well model.
-
getWellEquationNames(well, resmodel)¶ Get the names and types for the well equations of the model.
-
isInjector(well)¶ Check if well is an injector
Synopsis:
isInj = well.isInjector();
Parameters: well – Well class instance Returns: isInjector – boolean indicating if well is specified as an injector. Wells with sign zero is in this context defined as producers.
-
updateConnectionPressureDrop(well, wellSol0, wellSol, model, q_s, bhp, packed, dt, iteration)¶ Update the pressure drop within the well bore, according to a hydrostatic pressure distribution from the bottom-hole to the individual perforations.
To avoid dense linear systems, this update only happens at the start of each nonlinear loop.
-
updateConnectionPressureDropState(well, model, wellSol, rho_res, rho_well, mob_res)¶ Simpler version
-
updateLimits(well, wellSol0, wellSol, model, q_s, bhp, wellvars, p, mob, rho, dissolved, comp, dt, iteration)¶ Update solution variables and wellSol based on the well limits. If limits have been reached, this function will attempt to re-initialize the values and change the controls so that the next step keeps within the prescribed ranges.
-
updateWell(well, W)¶ Update well with a new control struct.
Synopsis:
well = well.updateWell(W);
Parameters: - model – Well model.
- W – Well struct representing the same wells, but with changed controls, active perforations and so on.
Returns: model – Updated well model.
-
updateWellSol(well, wellSol, variables, dx, resmodel)¶ Update function for the wellSol struct
-
updateWellSolAfterStep(well, resmodel, wellSol, wellSol0)¶ Updates the wellSol after a step is complete (book-keeping)
-
validateWellSol(well, resmodel, wsol, state)¶ #ok Validate wellSol for simulation
Synopsis:
wellSol = well.validateWellSol(model, wellSol, resSol)
Description:
Validate if wellSol is suitable for simulation. This function may modify the wellSol if the errors are fixable at runtime, otherwise it should throw an error. Note that this function is analogous to validateState in the base model class.
Parameters: - well – Well model class instance.
- model –
ReservoirModelclass instance. - wellSol – Well solution to be updated.
- resSol – Reservoir state
Returns: wellSol – Updated well solution struct.
-
VFPTable= None¶ Vertical lift table. EXPERIMENTAL.
-
allowControlSwitching= None¶ Limit reached results in well switching to another control
-
allowCrossflow= None¶ Boolean indicating if the well perforations allow cross-flow
-
allowSignChange= None¶ BHP-controlled well is allowed to switch between production and injection
-
dpMaxAbs= None¶ Maximum allowable absolute change in well pressure
-
dpMaxRel= None¶ Maximum allowable relative change in well pressure
-
dsMaxAbs= None¶ Maximum allowable change in well composition/saturation
-
-
class
UniformFacilityModel(varargin)¶ Bases:
FacilityModelSimplified facility model which is sometimes faster
-
combineMSwithRegularWells(W_regular, W_ms)¶ Combine regular and MS wells, accounting for missing fields
-
computeWellContributionsSingleWell(wellmodel, wellSol, resmodel, q_s, pBH, packed)¶ Main internal function for computing well equations and source terms
-
nozzleValve(v, rho, D, dischargeCoeff, flowtype)¶ Nozzle valve model
-
packPerforationProperties(W, p, mob, rho, dissolved, comp, wellvars, wellvarnames, varmaps, wellmap, ix)¶ Extract variables corresponding to a specific well
Synopsis:
packed = packPerforationProperties(W, p, mob, rho, dissolved, comp, wellvars, wellvarnames, varmaps, wellmap, ix)
Parameters: - W – Well struct for the specific well under consideration.
- p – Reservoir pressure for all cells in the domain.
- mob – Cell array with phase mobilities in each cell in the reservoir. Number of active phases long.
- rho – Cell array with phase densities in each cell in the reservoir. Number of active phases long.
- dissolved – Black-oil specific array of rs/rv. See the function ThreePhaseBlackOilModel>getDissolutionMatrix for details. Should be empty for models without dissolution ratios.
- comp – Cell array of components. Each entry should contain all reservoir cell values of that component, subject to whatever ordering is natural for the model itself.
- wellvars – Extended variable set added by the FacilityModel (see SimpleWell>getExtraPrimaryVariables)
- varmaps, wellmap, ix (wellvarnames,) – Internal book-keeping for
- added primary variables. (additional) –
Returns: packed – Struct where the values for the perforations of the well has been extracted.
NOTE: This function is intended for internal use in FacilityModel.
See also
FacilityModel
-
reorderWellPerforationsByDepth(W, active)¶ Undocumented Utility Function
-
selectFacilityFromDeck(deck, model)¶ Pick FacilityModel from input deck
-
setupMSWellEquationSingleWell(wm, model, wellSol0, wellSol, q_s, bhp, pN, alpha, vmix, resProps, dt, iteration)¶ Setup well residual equations for multi-segmented wells - and only those.
Synopsis:
function [eqs, eqsMS, cq_s, mix_s, status, cstatus, cq_r] = setupMSWellEquations(wm, ... model, wellSol0, dt, bhp, q_s, pN, vmix, alpha, wellNo)Parameters: - wm – Simulation well model.
- model – Resservoir Simulation model.
- wellSol0 – List of well solution structures from previous step
- dt – time step
- bhp – bottom hole pressure
- q_s – volumetric phase injection/production rate
- pN – pressure at nodes
- vmix – mixture mass flux in segments
- alpha – phase composition ratio at nodes
- wellNo – Well number identifying the well for which the equations are going to be assembled (should correspond to a multisegmented well)
Returns: eqs – Standard well equations: Cell array of mass balance equations at the connections - 1 equation for each phase,
eqsMS – multisegment well equations: Cell array of mass balance equations for each node and of pressure equations
- mass conservation equations for each phase, Dimension for each equation: number of nodes - 1
- pressure equation Dimension: number of segments
cq_s – List of vectors containing volumetric component source-terms (surface conds).
mix_s – List of vectors containing volumetric mixture of components in wellbores at connections (surface conds).
status – Logic vector of well statuses
cstatus – Logic vector of well connection statuses
cq_r – List of vectors containing volumetric phase source-terms (reservoir conds).
See also
FacilityModel
-
setupWellControlEquationsSingleWell(well, sol0, sol, pBH, q_s, status, mix_s, model)¶ Setup well controll (residual) equations
Synopsis:
eq = setupWellControlEquations(sol, pBH, q_s, status, mix_s, model)
Parameters: - sol – List of current well solution structures containing control type for each well
- pBH – Vector of well bhps
- q_s – List of vectors of well component volume-rates (surface conds)
- status – Logic vector of well statuses
- mix_s – List of vectors containing volumetric mixture of components in wellbroe at connections (surface conds).
- model – Simulation model.
Returns: eq – Well control equations
See also
WellModel, computeWellContributionsNew.
-
unpackPerforationProperties(packed)¶ Unpack the properties extracted by packPerforationProperties. Internal function.
-
wellBoreFriction(v, rho, mu, D, L, roughness, flowtype, assumeTurbulent)¶ Empricial model for well-bore friction
-
Contents¶ VFP
- Files
- VFPTable -
Simulators¶
-
Contents¶ SIMULATORS
- Files
- computeSensitivitiesAdjointAD - Compute parameter sensitivities using adjoint simulation computeGradientAdjointAD - Compute gradients using an adjoint/backward simulation that is linear in each step computeGradientPerturbationAD - Compute gradients using finite difference approximation by perturbing controls simulateScheduleAD - Run a schedule for a non-linear physical model using an automatic differention
-
computeGradientAdjointAD(state0, states, model, schedule, getObjective, varargin)¶ Compute gradients using an adjoint/backward simulation that is linear in each step
Synopsis:
grad = computeGradientAdjointAD(state0, states, model, schedule, getObjective
Description:
For a given schedule, compute gradients of objective with respect to well controls by solving adjoint equations.
Parameters: - state0 – Physical model state at t = 0
- states – All previous states. Must support the syntax
state = states{i}. If the problem is too large to fit in memory, it can use ResultHandler class to retrieve files from the disk. - model – Subclass of
PhysicalModelclass such asThreePhaseBlackOilModelthat models the physical effects we want to study. - schedule – Schedule suitable for
simulateScheduleAD. - getObjective – Function handle for getting objective function value for a given timestep with derivatives. Format: @(tstep)
Keyword Arguments: - ‘Verbose’ – Indicate if extra output is to be printed such as detailed convergence reports and so on.
- ‘scaling’ – Struct with fields
rateandpressureused to scale the relevant control equations, if the model supports it. - ‘LinearSolver’ – Subclass of
LinearSolverADsuitable for solving the adjoint systems.
Returns: gradients – Cell array of gradients for each control step.
-
computeGradientPerturbationAD(state0, model, schedule, getObjective, varargin)¶ Compute gradients using finite difference approximation by perturbing controls
Synopsis:
grad = computeGradientPerturbationAD(state0, model, schedule, getObjective)
Description:
For a given schedule, compute gradients with regards to well controls by perturbing all controls ever so slightly and re-running the simulation.
As the cost of this routine grows is approximately
(# wells)x(# ctrl step) x cost of schedule
it can be potentially extremely expensive. It is better to use the
computeGradientAdjointADroutine for most practical purposes. This routine is primarily designed for validation of said routine.Parameters: - state0 – Physical model state at
t = 0 - model – Subclass of
PhysicalModelclass such asThreePhaseBlackOilModelthat models the physical effects we want to study. - schedule – Schedule suitable for
simulateScheduleAD. - getObjective – Function handle for getting objective function value
from a set of wellSols.
Function handle format:
@(wellSols, states, schedule)
Keyword Arguments: - ‘Verbose’ – Indicate if extra output is to be printed such as detailed convergence reports and so on.
- ‘scaling’ – Struct with fields
rateandpressureused to scale the numerical perturbation. - ‘perturbation’ – Magnitude of perturbation. Default
1e-7.
Returns: grad – Gradients for each step.
See also
- state0 – Physical model state at
-
computeSensitivitiesAdjointAD(state0, states, model, schedule, getObjective, varargin)¶ Compute parameter sensitivities using adjoint simulation
Synopsis:
sens = computeSensitivitiesAdjointAD(state0, states, model, schedule, getObjective, varargin)
Description:
For a given schedule, compute senistivities with regards to parameters
Parameters: - state0 – Physical model state at
t = 0 - states – All previous states. Must support the syntax
state = states{i}. If the problem is too large to fit in memory, it can useResultHandlerclass to retrieve files from the disk. - model – Subclass of PhysicalModel class such as
ThreePhaseBlackOilModelthat models the physical effects we want to study. - schedule – Schedule suitable for
simulateScheduleAD. - getObjective – Function handle for getting objective function value for a given timestep with derivatives. Format: @(tstep)
Keyword Arguments: - ‘Parameters’ – Defaults to
{'transmissibility', 'porevolumes'}. - ‘ParameterTypes’ – Defaults to ‘value’ for all. The other option is ‘multiplier’
- ‘Regularization’ – One (sparse) matrix for each parameter-set, requires also modelBase
- ‘LinearSolver’ – Subclass of
LinearSolverADsuitable for solving the adjoint systems.
Returns: sens – Sensitivites.
See also
computeGradientAD- state0 – Physical model state at
-
simulateScheduleAD(initState, model, schedule, varargin)¶ Run a schedule for a non-linear physical model using an automatic differention
Synopsis:
wellSols = simulateScheduleAD(initState, model, schedule) [wellSols, state, report] = simulateScheduleAD(initState, model, schedule)
Description:
This function takes in a valid schedule file (see required parameters) and runs a simulation through all timesteps for a given model and initial state.
simulateScheduleAD is the outer bookkeeping routine used for running simulations with non-trivial control changes. It relies on the model and (non)linear solver classes to do the heavy lifting.
Parameters: - initState – Initial reservoir/model state. It should have whatever fields are associated with the physical model, with reasonable values. It is the responsibility of the user to ensure that the state is properly initialized.
- model – The physical model that determines jacobians/convergence
for the problem. This must be a subclass of the
PhysicalModelbase class. - schedule –
Schedule containing fields step and control, defined as follows:
schedule.controlis a struct array containing fields that the model knows how to process. Typically, this will be the fields such asWfor wells orbcfor boundary conditions.schedule.stepcontains two arrays of equal size namedvalandcontrol. Control is a index into theschedule.controlarray, indicating which control is to be used for the timestep.`schedule.step.val` is the timestep used for that control step.
Keyword Arguments: ‘Verbose’ – Indicate if extra output is to be printed such as detailed convergence reports and so on.
‘OutputMinisteps’ – The solver may not use timesteps equal to the control steps depending on problem stiffness and timestep selection. Enabling this option will make the solver output the states and reports for all steps actually taken and not just at the control steps. See also
convertReportToSchedulewhich can be used to construct a new schedule from these timesteps.‘initialGuess’ – A cell array with one entry per control-step. If provided, the state from this cell array is passed as initial guess for the
NonLinearSolver. See:NonLinearSolver.solveTimestep, optional arguments.‘NonLinearSolver’ – An instance of the
NonLinearSolverclass. Consider using this if you for example want a special timestep selection algorithm. See theNonLinearSolverclass docs for more information.‘OutputHandler’ – Output handler class, for example for writing states to disk during the simulation or in-situ visualization. See the ResultHandler base class.
‘WellOutputHandler’ – Same as ‘OutputHandler’, but for the well solutions for the individual report steps. Well solutions are also stored using OutputHandler, but using WellOutputHandler is convenient for quickly loading well solutions only.
‘ReportHandler’ – Same as ‘OutputHandler’, but for the reports for the individual report steps.
‘LinearSolver’ – Class instance subclassed from
LinearSolverAD. Used to solve linearized problems in theNonLinearSolverclass. Note that if you are passing aNonLinearSolver, you might as well put it there.‘afterStepFn’ – Function handle to an optional function that will be called after each successful report step in the schedule. The function should take in the following input arguments: - model: The model used in the schedule - states: A cell array of all states that are
computed, as well as possible empty entries where the states have not been computed yet.
- reports: A cell array of reports for each step, with empty entries for steps that have not been reached yet.
- solver: The NonLinearSolver instance.
- schedule: The current schedule.
- simtime: Array with the time in seconds taken by
the
NonLinearSolverto compute each step. Entries not computed will contain zeros.
See
getPlotAfterStepfor more information andhowtoAddPlotHookfor a worked example.‘controlLogicFn’ – Function handle to optional function that will be called after each step enabling schedule updates to be triggered on specified events. Input arguemnts: - state: The current state - schedule: The current schedule - report: Current report - i: The current report step such that current
control step equals schedule.step.control(i)
The function must have three outputs: - schedule: Possibly updated schedule - report: Possibly updated report - isAltered: Flag indicated whether the schedule
was updated or not
Returns: - wellSols – Well solution at each control step (or timestep if ‘OutputMinisteps’ is enabled.
- states – State at each control step (or timestep if ‘OutputMinisteps’ is enabled.
- schedulereport – Report for the simulation. Contains detailed info for the whole schedule run, as well as arrays containing reports for the whole stack of routines called during the simulation.
Note
For examples of valid models, see
ThreePhaseBlackOilModelorTwoPhaseOilWaterModel.See also
computeGradientAdjointAD,PhysicalModel
Solvers¶
-
Contents¶ SOLVERS
- Files
- AMGCL_CPRSolverAD - Linear solver that calls external compiled multigrid solver AGMGSolverAD - Linear solver that calls external compiled multigrid solver AMGCLSolverAD - Linear solver that calls external compiled multigrid solver BackslashSolverAD - Linear solver that calls standard MATLAB direct solver mldivide “” CPRSolverAD - Solve a problem with a pressure component using constrained a pressure residual method getNonLinearSolver - Set up reasonable defaults for the nonlinear solver for a field GMRES_ILUSolverAD - Preconditioned GMRES solver. HandleLinearSolverAD - Simple solver for wrapping functions on the form x = fn(A, b); LinearizedProblem - A linearized problem within a non-linear iteration LinearSolverAD - Base class for linear solvers in the AD framework NonLinearSolver - Generalized Newton-like nonlinear solver NoOpSolverAD - Linear solver that does nothing.
-
class
AGMGSolverAD(varargin)¶ Linear solver that calls external compiled multigrid solver
Synopsis:
solver = AGMGSolverAD()
Description:
This solver calls the AGMG package and supports setup/cleanup of the multigrid solver for multiple solves of the same problem.
Note
This solver requires AGMG to be installed and working.
See also
BackslashSolverAD-
cleanupSolver(solver, A, b, varargin)¶ Clean up solver after use (if needed)
-
setupSolver(solver, A, b, varargin)¶ Run setup on a solver for a given system
-
reuseSetup= None¶ Will reuse the setup phase to improve speed for e.g. a GMRES loop with the same matrix system. However, some systems report segfaults with this option enabled.
-
setupDone= None¶ Internal book-keeping variable
-
-
class
AMGCLSolverAD(varargin)¶ Linear solver that calls external compiled multigrid solver
Synopsis:
solver = AMGCLSolverAD()
Description:
AD-interface for the AMGCL interface.
Note
This solver requires AMGCL to be installed and working.
See also
BackslashSolverAD-
cleanupSolver(solver, A, b, varargin)¶ #ok
-
amgcl_setup= u'1'¶ 1 for no reuse, 2 for re-use
-
-
class
AMGCL_CPRSolverAD(varargin)¶ Linear solver that calls external compiled multigrid solver
Synopsis:
solver = AMGCLSolverAD()
Description:
AD-interface for the AMGCL interface.
Note
This solver requires AMGCL to be installed and working.
See also
BackslashSolverAD-
getDiagonalInverse(solver, A)¶ Reciprocal of diagonal matrix
-
undoScaling(solver, x, scaling)¶ #ok Undo effects of scaling applied to linear system
-
couplingTol= u'0.02'¶ tolerance for drs
-
decoupling= u"'trueIMPES'"¶ trueimpes, quasiimpes, none
-
diagonalTol= u'0.2'¶ tolerance if strategy ends with _drs
-
doApplyScalingCPR= None¶ true / false
-
pressureScaling= None¶ scaling factor for pressure - automatically determined
-
strategy= u"'mrst'"¶ mrst, mrst_drs, amgcl, amgcl_drs
-
useSYMRCMOrdering= None¶ true / false
-
-
class
BackslashSolverAD(varargin)¶ Linear solver that calls standard MATLAB direct solver mldivide “”
Synopsis:
solver = BackslashSolverAD()
Description:
This solver solves linearized problems using matlab builtin mldivide.
See also
LinearSolverAD
-
class
CPRSolverAD(varargin)¶ Solve a problem with a pressure component using constrained a pressure residual method
Synopsis:
solver = CPRSolverAD()
Description:
Solve a linearized problem with a significant elliptic/pressure component via a two stage preconditioner for GMRES. By exposing the elliptic component as a seperate system, a special elliptic solver can be used to handle the highly connected components.
For second stage, ILU(0) is used.
Keyword Arguments: ‘property’ – Set property to value. See also
BackslashSolverAD,LinearSolverAD,LinearizedProblem-
solveLinearProblem(solver, problem0, model)¶ Solve a linearized problem using a constrained pressure residual preconditioner
-
solveLinearSystem(solver, A, b)¶ #ok
-
diagonalTol= None¶ Diagonal tolerance in [0,1].
-
ellipticSolver= None¶ LinearSolverAD subclass suitable for the elliptic submatrix.
-
ellipticVarName= None¶ Name of elliptic-like variable which will be solved using elliptic solver
-
pressureScaling= None¶ Scaling factor applied to pressure equations
-
relativeTolerance= None¶ Relative tolerance for elliptic solver
-
trueIMPES= None¶ Use true impes decoupling strategy (if supported by model)
-
-
class
GMRES_ILUSolverAD(varargin)¶ Preconditioned GMRES solver.
Synopsis:
solver = GMRES_ILUSolverAD()
Description:
Solve a linearized problem using a GMRES solver with ILU preconditioner.
Parameters: None – Keyword Arguments: ‘property’ – Set property to value. See also
BackslashSolverAD,CPRSolverAD,LinearizedProblem,LinearSolverAD-
dropTolerance= None¶ Modified ilu type: ‘row’, ‘col’, {‘off’}
-
ilutype= None¶ Drop tolerance for elements (default: 0)
-
modifiedIncompleteILU= None¶ Boolean indicating replacement of zero diagonal entries
-
pivotThreshold= None¶ Reorder equations when diagonal entries are zero for ilu0
-
udiagReplacement= None¶ Pivoting threshold for ilupt. Default 1.
-
-
class
HandleLinearSolverAD(fcn)¶ Simple solver for wrapping functions on the form x = fn(A, b);
-
getDescription(solver)¶ Get the description and a short name used for display purposes.
-
solveLinearSystem(solver, A, b)¶ #ok
-
-
class
LinearSolverAD(varargin)¶ Base class for linear solvers in the AD framework
Synopsis:
solver = LinearSolverAD()
Description:
Base class for linear solvers. Implement methods for solving linearized problems and adjoints. Also supports setup/cleanup functions before/after use for initialize once-type usage.
Parameters: None – Keyword Arguments: ‘property’ – Set property to value. Note
This class is intended as superclass. It cannot actually solve problems.
See also
BackslashSolverAD,CPRSolverAD,LinearizedProblem-
getSolveReport(solver, varargin)¶ #ok
-
solveAdjointProblem(solver, problemPrev, problemCurr, adjVec, objective, model, varargin)¶ #ok
-
solveLinearSystem(solver, A, b)¶ #ok Solve the linear system to a given tolerance
-
applyLeftDiagonalScaling= None¶ Apply left diagonal scaling before solving
-
applyRightDiagonalScaling= None¶ Apply right diagonal scaling before solving
-
equationOrdering= None¶ Equation ordering to be used for linear solver. Row vector of equal length to the size of the linear system.
-
extraReport= None¶ Enable this to produce additional report output
-
id= u"''"¶ Short text string identifying the specific solver. Appended to the short name (see getDescription)
-
keepNumber= None¶ If set, linear solver will reduce the system to the first keepNumber entries
-
maxIterations= None¶ Max number of iterations used
-
reduceToCell= None¶ Reduce to per-cell system before solving
-
replaceInf= None¶ Boolean indicating if the solver should replace Inf in the results
-
replaceNaN= None¶ Boolean indicating if the solver should replace NaN in the results
-
replacementInf= None¶ If replaceInf is enabled, this is the value that will be inserted
-
replacementNaN= None¶ If replaceNaN is enabled, this is the value that will be inserted
-
tolerance= None¶ Linear solver tolerance
-
useSparseReduction= None¶ If true, sparse indexing will be used with keepNumber option
-
variableOrdering= None¶ Variable ordering to be used for linear solver. Row vector of equal length to the size of the linear system.
-
verbose= None¶ Verbose output enabler
-
-
class
LinearizedProblem(varargin)¶ A linearized problem within a non-linear iteration
Synopsis:
problem = LinearizedProblem(primvars, state) problem = LinearizedProblem(primvars, state, dt) problem = LinearizedProblem(eqs, eqtypes, eqnames, primvars, state) problem = LinearizedProblem(eqs, eqtypes, eqnames, primvars, state, dt)
Description:
A class that implements storage of an instance of a linearized problem discretized using AD. This class contains the residual equations evaluated for a single state along with meta-information about the equations and the primary variables they are differentiated with respect to.
A linearized problem can be transformed into a linear system and solved using
LinearSolverAD-derived subclasses, given that the number of equations matches the number of primary variables.In particular, the class contains member functions for:
- assembling a linear system from the Jacobian block matrices stored for each individual (continuous) equation
- appending/prepending additional equations
- using a block-Gaussian method to eliminate individual variables or all variables that are not of a specified type
- recovering increments corresponding to variables that have previously been eliminated
- computing the norm of each residual equation
as well as a number of utility functions for sanity checks, quering of indices and the number of equations, clearing the linear system, etc.
See also
LinearSolverAD,PhysicalModel-
appendEquations(problem, equations, types, names)¶ Add one or more equations to the beginning of the current list of equations.
-
assembleSystem(problem)¶ Assemble the linear system from the individual Jacobians and residual functions. Note the return parameter, as problem is not a handle class.
-
checkInputs(problem, equations, types, names)¶ #ok
-
clearSystem(problem)¶ Remove pre-assembled linear system. Typically used if the equations are manually changed, and one does not want to evaluate inconsistent linear systems.
-
countOfType(problem, name)¶ Count the number of equations that are of a specific type.
-
eliminateVariable(problem, variable)¶ Eliminate a variable from the problem using the equation with the same index.
Parameters: variable – The name of the variable (corresponding to an entry in problem.names) that is to be eliminated.
Returns: - problem – Modified problem.
- eliminatedEquation – The equation that was eliminated.
Note
For non-diagonal matrices, the cost of the equation elimination can be large.
-
getEquationVarNum(problem, n)¶ Get number of subequations for one or more equations. A single equation is a collection of a number of equations grouped by the constructor. Typically, all subequations should be of the same :param n: (OPTIONAL) Indices of the equations for which the number
of subequations is desired. If omitted, the function returns the number for all equations.Returns: varnum – Array with the number of subequations for the requested equations.
-
getLinearSystem(problem)¶ Get the linear system from the individual Jacobians and residual equations in a format suitable for general linear solvers. If the equations are not already assembled, this will result in a call to assembleSystem. If you will retrieve the linear system multiple times, it is better to call assembleSystem beforehand.
-
indexOfEquationName(problem, name)¶ Get the index into the list of equations for a specific name
-
indexOfPrimaryVariable(problem, name)¶ Get the index of a primary variable by name.
-
indexOfType(problem, name)¶ Get the index(es) of a type of variable by name.
-
norm(problem, varargin)¶ Get the norm of each equation. This is an overloaded function.
-
numel(problem)¶ Get the number of distinct equations. Note that an equation here can refer to multiple subequations of the same type (e.g. a 100 cell problem where the equations are oil and water conservation laws will have two equations in total, with each having 100 sub-entries.
-
prependEquations(problem, equations, types, names)¶ Add one or more equations to the end of the current list of equations.
-
processResultAfterSolve(problem, result, report)¶ #ok Do nothing
-
recoverFromSingleVariableType(reducedProblem, originalProblem, incrementsReduced, eliminated)¶ Recover the increments in primary variables corresponding to equations that have previously been eliminated by for instance “reduceToSingleVariableType”. :param originalProblem: The problem before elimination. :param incrementsReduced: Increments from the solution of the reduced
problem.Parameters: eliminated – The eliminated equations. Returns: dx – All increments, as if the originalProblemwas solved directly.
-
reduceToSingleVariableType(problem, type)¶ Eliminate all equations that are not of a given type
Parameters: type – String matching one of problem.types. Equations that DO NOT have that type will be eliminated.
Returns: - problem – Modified problem.
- eliminated – Eliminated equations. Can be used to recover the eliminated values later.
Note
Can have a severe cost depending on the sparsity patterns involved in the different equations. Typical usage is to eliminate non-cell equations (wells, control equations).
-
reorderEquations(problem, newIndices)¶ Reorder equations based on a set of indices. ARGUMENTS: - newIndices: numel(problem) long array of new indices into the
equations.OUTPUT: - problem : Problem with re-numbered equations.
-
A= None¶ Linear system after assembling Jacobians.
-
b= None¶ Right hand side for linearized system. See A.
-
drivingForces= None¶ The driving forces struct used to generate the equations.
-
dt= None¶ The time step length (Not relevant for all problems)
-
equationNames= None¶ Cell array of equal length to number of equations, giving them unique names
-
equations= None¶ Cell array of the equations. Can be doubles, or more typically ADI objects.
-
iterationNo= None¶ Nonlinear iteration number corresponding to problem.
-
primaryVariables= None¶ Cell array containing the names of the primary variables.
-
state= None¶ The problem state used to produce the equations
-
types= None¶ Cell array of equal length to number of equations, with strings indicating their types
-
class
NoOpSolverAD¶ Linear solver that does nothing.
Synopsis:
solver = NoOpSolverAD()
Description:
Debug solver. It has the correct interfaces, but it always returns zero as the solution for the problem. It is, however, very fast…
Note
You should not use this solver.
See also
BackslashSolverAD
-
class
NonLinearSolver(varargin)¶ Generalized Newton-like nonlinear solver
Synopsis:
solver = NonLinearSolver() solver = NonLinearSolver('maxIterations', 5)
Description:
The NonLinearSolver class is a general non-linear solver based on Newton’s method. It is capable of timestep selection and cutting based on convergence rates and can be extended via subclassing or modular linear solvers and timestep classes.
Convergence is handled by the PhysicalModel class. The NonLinearSolver simply responds based on what the model reports in terms of convergence to ensure some level of encapsulation.
Parameters: None. – Returns: A NonLinearSolver class instance ready for use. See also
simulateScheduleAD,LinearSolverAD,SimpleTimeStepSelector-
checkForOscillations(solver, res, index)¶ #ok Check if residuals are oscillating. They are oscillating of the ratio of forward and backwards differences for a specific residual is negative.
-
checkForStagnation(solver, res, index)¶ Check if residuals have stagnated. Residuals are flagged as stagnating if the relative change is smaller than the tolerance (in absolute value).
-
solveMinistep(solver, model, state, state0, dt, drivingForces)¶ Attempt to solve a single mini timestep while trying to avoid stagnation or oscillating residuals.
-
solveTimestep(solver, state0, dT, model, varargin)¶ Solve a timestep for a non-linear system using one or more substeps :param state0: State at the beginning of the timestep :param dT: Timestep size. The solver will move forwards
either as a single step or multiple substeps depending on convergence rates and sub timestep selection.Parameters: model – Model inheriting from PhysicalModel with a valid implementation of the “stepFunction” member function.
Keyword Arguments: ‘W’ – Wells for the timestep. (struct)
‘bc’ – Boundary conditions for the problem (struct).
‘src’ – Source terms for the timestep (struct).
NOTE – Wells, boundary conditions and source terms are the standard types of external forces in MRST. However, the model input determines which of these are actually implemented for that specific step function. Not all combinations are meaningful for all models.
Some models may implement other types of external forces that have other names, specified in the model’s “getValidDrivingForces” method.
Returns: - state – Problem state after timestep, i.e. if state0 held pressure, saturations, … at T_0, state now holds the same values at T_0 + dT.
- report – Report struct, containing some standard information (iteration count, convergence status etc) in addition to any reports the stepFunction contains.
- ministates – Cell array containing all ministeps used to get to T = T_0 + dt. If the solver decided to take a single step and was successful, this will just be {state}.
See also
PhysicalModel
-
stabilizeNewtonIncrements(solver, model, problem, dx)¶ #ok<INUSL> Attempt to stabilize newton increment by changing the values of the increments.
-
LinearSolver= None¶ The solver used to solve the linearized problems during the simulation.
-
acceptanceFactor= u'1'¶ If we run out of iterations, this factor used for a relaxed tolerance check.
-
alwaysUseLinesearch= u'false'¶ Debug option to always use line search
-
continueOnFailure= u'false'¶ Continue even if failure is reported by the model. Results are most likely not useful. Intended for nested nonlinear solvers.
-
enforceResidualDecrease= u'false'¶ Abort a solution if no reduction is residual is happening.
-
errorOnFailure= u'true'¶ If error on failure is not enabled, the solver will return even though it did not converge. May be useful for debugging. Results should not be relied upon if this is enabled. If errorOnFailure is disabled, the solver will continue after a failed timestep, treating it as a simply non-converged result with the maximum number of iterations
-
identifier= u"''"¶ String identifier for the nonlinear solver
-
linesearchConvergenceNames= u'{}'¶ Residual names to be checked in linesearch
-
linesearchDecreaseFactor= u'1'¶ Required reduction factor in residual (default 1)
-
linesearchMaxIterations= u'10'¶ Max iterations in linesearch
-
linesearchReductionFactor= u'1/2'¶ Reduction factor for each step in LS
-
linesearchReductionFn= u'[]'¶ Function for combining multiple residuals into value used by linesearch
-
linesearchResidualScaling= u'[]'¶ Residual scaling for each equation
-
maxIterations= u'25'¶ The max number of iterations during a ministep.
-
maxRelaxation= u'1.0'¶ Largest possible relaxation factor
-
maxTimestepCuts= u'6'¶ The maximum number of times the timestep can be halved before it is counted as a failed attempt
-
minIterations= u'1'¶ The minimum number of solves during a ministep.
-
minRelaxation= u'0.5'¶ Lowest possible relaxation factor
-
relaxationDecrement= u'[]'¶ Change in relaxation on stagnation/oscillation
-
relaxationIncrement= u'0.1'¶ Change in relaxation on stagnation/oscillation
-
relaxationParameter= u'1'¶ Relaxation parameter between 0 and 1.
-
relaxationType= u"'dampen'"¶ Relaxation is reduced by this when stagnation occurs
-
reportLevel= u'0'¶ Amount of data to be output in report. Higher numbers may give more output.
-
stagnateTol= u'1e-2'¶ Stagnation tolerance - used in relaxation to determine of a residual value is no longer decreasing
-
timeStepSelector= None¶ Subclass of SimpleTimeStepSelector used to select timesteps
-
useLinesearch= u'false'¶ True to enable line-search in residual
-
useRelaxation= u'false'¶ Boolean indicating if Newton increments should be relaxed.
-
verbose= u'[]'¶ Verbose flag used to get extra output during simulation.
-
-
getNonLinearSolver(model, varargin)¶ Set up reasonable defaults for the nonlinear solver for a field simulation with significant size and complexity
Synopsis:
solver = getNonLinearSolver(model)
Parameters: model – Simulation model (subclass of PhysicalModel).
Keyword Arguments: - ‘DynamicTimesteps’ – Set up a simple iteration count timestep selector.
- ‘useCPR’ – Set up CPR-type preconditioner as the linear solver. Will try to use the best known linear solver (either AGMG or Matlab builtin at the moment).
Returns: solver – NonLinearSolver instance.
See also
simulateScheduleAD, NonLinearSolver
Time-step selection¶
-
Contents¶ TIMESTEPS
- Files
- IterationCountTimeStepSelector - Adjust timesteps based with target iteration count, based on history rampupTimesteps - Create timesteps that ramp up geometrically SimpleTimeStepSelector - Time step selector base class StateChangeTimeStepSelector - The StateChangeTimeStepSelector is a time-step selector that attempts to
-
class
IterationCountTimeStepSelector(varargin)¶ Adjust timesteps based with target iteration count, based on history
Synopsis:
selector = IterationCountTimeStepSelector(); selector = IterationCountTimeStepSelector('targetIterationCount', 5);
Description:
Routine used for dynamic timestepping
Parameters: None. –
Keyword Arguments: - targetIterationCount – Desired number of iterations.
- iterationOffset – Uses [actual + iterationOffset] to calculate the parameter. Larger values makes the step selector less aggressive for iteration targets near zero.
- (Other options) – Inherited from SimpleTimeStepSelector.
Returns: Time step selector.
See also
SimpleTimeStepSelector, NonLinearSolver
-
computeTimestep(selector, dt, dt_prev, model, solver, state_prev, state_curr, forces)¶ Dynamically compute timestep
-
iterationOffset= None¶ Offset to make iteration a bit smoother as a response function.
-
targetIterationCount= None¶ Desired number of nonlinear iterations per timestep.
-
class
SimpleTimeStepSelector(varargin)¶ Time step selector base class
Synopsis:
selector = SimpleTimeStepSelector(); selector = SimpleTimeStepSelector('maxTimestep', 5*day);
Description:
The timestep selector base class is called by the NonLinearSolver to determine timesteps, based on hard limits such as the min/max timesteps as well as possibly more advanced features via the computeTimestep method that can account for iteration count, residual reduction etc.
Parameters: None – Keyword Arguments: See properties Returns: selector suitable for passing to the NonLinearSolver class. See also
IterationCountTimeStepSelector, NonLinearSolver, StateChangeTimeStepSelector
-
computeTimestep(selector, dt, dt_prev, model, solver, state_prev, state_curr, forces)¶ #ok Compute timestep dynamically - does nothing for base class
-
newControlStep(selector, control)¶ Determine if controls have changed
-
controlsChanged= None¶ Flag indicating that the controls have changed
-
firstRampupStep= None¶ The first ministep attempted after controls have changed
-
firstRampupStepRelative= None¶ Relative version of firstRampupStep
-
history= None¶ Stored history used to pick next timestep
-
isFirstStep= None¶ Flag indicating that we are at the start of the simulation
-
isStartOfCtrlStep= None¶ Flag indicating the beginning of a control step
-
maxHistoryLength= None¶ The maximum number of history steps stored
-
maxRelativeAdjustment= None¶ Ensure dt_next < dt_suggested*maxRelativeAdjustment
-
maxTimestep= None¶ Hard upper limit on timestep in seconds
-
minRelativeAdjustment= None¶ Ensure dt_next > dt_suggested*minRelativeAdjustment
-
minTimestep= None¶ Hard lower limit on timestep in seconds
-
previousControl= None¶ Previous control seen by the selector used to determine when controls change.
-
resetOnControlsChanged= None¶ Reset when controls change
-
stepLimitedByHardLimits= None¶ Flag indicating that hard limits and not any step algorithm was the cause of the previous timestep taken
-
verbose= None¶ Extra output
-
-
class
StateChangeTimeStepSelector(varargin)¶ The StateChangeTimeStepSelector is a time-step selector that attempts to ensure that certain properties of the state change at target rates during the simulation. This can often be a good way of controlling timesteps and minimizing numerical error if good estimates of the error are known.
See the doctoral dissertation of H. Cao “Development of techniques for general purpose simulators”, Stanford, 2002.
-
relaxFactor= u'1'¶ Relaxation factor. Larger values mean less severe changes in time-steps.
-
targetChangeAbs= u'[]'¶ Target change (absolute units). Double array of the same length as targetProps.
-
targetChangeRel= u'[]'¶ Target change (relative units). This is a double array of the
-
targetProps= u'{}'¶ The target properties to time control. Cell array of N strings,
-
-
rampupTimesteps(time, dt, n)¶ Create timesteps that ramp up geometrically
Synopsis:
dT = rampupTimesteps(1*year, 30*day) dT = rampupTimesteps(1*year, 30*day, 5)
Description:
This function generates a timestep sequence for a given total time interval that increases geometrically until it reaches some target timestep. The rest of the interval is then divided into a number of target timesteps.
Parameters: - time – The total simulation time so that sum(dt) = time
- dt – Target timestep after initial ramp-up
- n – (OPTIONAL) Number of rampup steps. Defaults to 8.
Returns: dt – Array of timesteps.
Note
The final timestep may be shorter than dt in order to exactly reach T.
Model upscaling¶
-
Contents¶ UPSCALE
- Files
- upscaleModelTPFA - Upscale a fine model into a coarser version using a partition vector upscaleSchedule - Upscale a schedule to a coarser model upscaleState - Create a upscaled state by simple processing of values
-
upscaleModelTPFA(model, partition, varargin)¶ Upscale a fine model into a coarser version using a partition vector
Synopsis:
modelcoarse = upscaleModelTPFA(model, partition)
Description:
Given a fine model and a partition vector (created using standard coarsegrid tools such as partitionUI), this routine makes a coarser model with upscaled properties. Using somewhat reasonable defaults, most parts of the routine can be overriden by better values if the user already knows for instance transmissibilities.
Parameters: - model – Fine scale model. Subclass of
ReservoirModel. - partition – Partition vector. Length equal to
model.G.cells.num, with positive indicator values. All cells with the same indicator will be combined into a single coarse block.
Keyword Arguments: - ‘validatePartition’ – Ensure partition is connected on the grid, and numbered from 1…N without gaps.
- ‘transCoarse’ – Coarse transmissibilities. Will be calculated from upscaled permeability if not provided.
- ‘transFromRock’ – Compute transmissibility from rock. Default is true. If disabled, the coarse transmissibility will be a sum instead.
- ‘permCoarse’ – Coarse permeability. Will be calculated using harmonic averaging if not provided.
- ‘neighborship’ – Coarse neighborship (matching transCoarse). Will be derived from fine grid if not provided.
- ‘poroCoarse’ – Coarse porosities. Computed from fine model using a simple sum if not provided.
Returns: model – Coarse model.
See also
- model – Fine scale model. Subclass of
-
upscaleSchedule(model, schedule, varargin)¶ Upscale a schedule to a coarser model
Synopsis:
schedule = upscaleSchedule(model, schedule)
Parameters: - model – The coarse model the schedule is to be converted to. Assumed to be derived from the fine model used with schedule.
- schedule – Schedule to be upscaled.
Keyword Arguments: ‘wellUpscaleMethod’ – Upscaling method for well indices. The default is to recompute the well indices in the new block. Other options are ‘sum’, ‘harmonic’ and ‘mean’. We recommend applying a dedicated upscaling routine and replacing these values if well-bore flow performance is important.
‘bcUpscaleMethod’ – Interpolation strategy used for boundary conditions. Possible options:
- linear: Default.
- idw: Inverse distance weighting
- mean: Mean value.
- nearest: Nearest neighbor.
In addition, any unknown arguments will be passed onto the interpolation routine used. Depending on the dimensionality and the boundary conditions, this is either
interp1orscatteredInterp.
Returns: schedule – Schedule upscaled for the coarse model.
Note
Support for boundary conditions relies on interpolation. Results should be examined before use for complex grids.
See also
-
upscaleState(coarsemodel, model, state)¶ Create a upscaled state by simple processing of values
Synopsis:
state_coarse = upscaleState(coarsemodel, model, state_fine)
Description:
Convert a state for a fine model into a realization of the same state for a coarse model.
Parameters: - coarsemodel – A coarse model derived from the fine model.
- model – The fine model. Subclass of
ReservoirModel. - state – State to be converted. Should correspond to
model.
Returns: state – Coarse state suitable for the coarsemodel.
See also
Plotting¶
-
Contents¶ PLOTTING
- Files
- getPlotAfterStep - Get a function that allows for dynamic plotting in
simulateScheduleAD. inspectFluidModel - Create a interactive plotting panel for a given model that shows plotReportIterations - Plot nonlinear convergence behavior for simulateScheduleAD output plotWellSols - Plot well solutions from AD-solvers simpleUIbar - Undocumented Utility Function simulationRuntimePanel - Internal function for drawing panel during simulation. See
-
getPlotAfterStep(state0, model, schedule, varargin)¶ Get a function that allows for dynamic plotting in
simulateScheduleAD.Synopsis:
fn = getPlotAfterStep(state0, model, schedule, 'plotWell', true);
Description:
The
simulateScheduleADfunction has a optional input argumentafterStepFnthat allows for dynamic plotting after each step in the simulation, for instance to show how the well curves progress during the simulation, or to print out extra information to the command window. This function is an implementation of one such function, that can add both a panel showing the simulation progress, as well as interactive plots for well and reservoir quantities.Parameters: - state0 – Initial state for simulateScheduleAD
- model – Simulation model which will be passed to simulateScheduleAD.
- schedule – The simulation schedule containing wells, driving forces and time-steps that will be passed to simulateScheduleAD.
Keyword Arguments: - ‘plotWell’ – Launch interactive plotting for well quantities
using
plotWellSols - ‘plotReservoir’ – Add an interactive plotting window for reservoir quantities during the simulation. Note that, due to limitations in the implementation, this window will only be truly interactive after the simulation finishes. You can, however, set the options (field for plotting, locked color axis and so on) before initiating the simulation itself.
- ‘view’ – View angle for the reservoir plotting. See Matlab
builtin
viewfor more information. Defaults to empty for no modification to the default. - ‘wells’ – Wells for the reservoir plotting (using
plotWell)
Returns: fn – Function handle suitable for the
afterStepFninput insimulateScheduleAD.Example:
fn = getPlotAfterStep(state0, model, schedule, 'plotWell', true); simulateScheduleAD(state0, model, schedule, 'afterStepFn', fn);
See also
-
inspectFluidModel(model, varargin)¶ Create a interactive plotting panel for a given model that shows different fluids properties.
Synopsis:
h = inspectFluidModel(model)
Description:
Launch an interactive plotting interface for the fluid model.
Parameters: model – Some ReservoirModel-derived class with a valid fluid model. This function has primarily been tested for black-oil and black-oil similar fluids.Keyword Arguments: ‘pressureRange’ – An array of the pressures values to be used for pressure-dependent properties. Defaults to max(model.minimumPressure, 0.1*barsa)tomin(model.maximumPressure, 600*barsa)Returns: h – Figure handle to the plotting panel.
-
plotReportIterations(report, schedule)¶ Plot nonlinear convergence behavior for simulateScheduleAD output
Synopsis:
plotReportIterations(report, schedule)
Description:
Creates a plot in the current figure of the nonlinear convergence behavior of a report and schedule pair. Timesteps and ministeps are visualized, along with green and red boxes that indicate successful and wasted iterations respectively.
Parameters: - report – Report as returned by
simulateScheduleAD. - schedule – The schedule passed to
simulateScheduleADto produce the report.
Returns: Nothing.
- report – Report as returned by
-
plotWellSols(wellsols, varargin)¶ Plot well solutions from AD-solvers
Synopsis:
plotWellSols(wellSols, time); plotWellSols(wellSols);
Description:
Open interactive plotting interface for well solutions.
Parameters: - wellSols – Cell array of
nstepby 1, each containing a uniform struct array of well solution structures. For example, the first output fromsimulateScheduleAD. Can also be a cell array of such cell arrays, for comparing multiple simulation scenarios. - time – (OPTIONAL) The time for each timestep. If not provided, the plotter will use step number as the x axis intead. If wellSols is a cell array of multiple datasets, time should also be a cell array, provided not all datasets use the same timesteps.
Keyword Arguments: - ‘field’ – Initial field for plotting (default: ‘bhp’).
- ‘linestyles’ – Cell array of line styles used for different datasets.
- ‘markerstyles’ – Marker array of line styles used for different datasets.
- ‘datasetnames’ – A cell array of dataset names used for the legend when plotting multiple datasets.
- ‘timescale’ – A string for the default choice for axis time-scale. A string which matches either choice: ‘days’, ‘minutes’, ‘seconds’, ‘hours’, ‘years’
Returns: - fh – figure handle to plotting panel
- inject – function handle used to dynamically inject new datasets into the viewer (for example, from a running simulation). Same syntax as the base function, but does not support additional varargin.
See also
- wellSols – Cell array of
-
simpleUIbar(parent, data, start, height, txt, varargin)¶ Undocumented Utility Function
-
simulationRuntimePanel(model, states, ctrl_reports, solver, schedule, simtime, varargin)¶ Internal function for drawing panel during simulation. See getPlotAfterStep.
Automatic differentation backends¶
-
Contents¶ BACKENDS
- Files
- AutoDiffBackend - Automatic differentiation backend class
-
class
AutoDiffBackend¶ Automatic differentiation backend class
Synopsis:
backend = AutoDiffBackend()
Description:
Base class for automatic differentiation backends. A backend is an implementation of automatic differentation and must support the initialization of one (or more) variables as AD objects.
Returns: Backend – Initialized class instance See also
DiagonalAutoDiffBackend,SparseAutoDiffBackend-
AutoDiffBackend()¶ Class constructor.
-
convertToAD(backend, v, sample)¶ Given a
sampleAD object, convertvto AD.
-
getBackendDescription(backend)¶ Get a text string describing the backend
-
initVariablesAD(backend, varargin)¶ Initialize variables as AD
-
updateDiscreteOperators(backend, model)¶ Modify model/operators for use with backend
Synopsis:
model = backend.updateDiscreteOperators(model)
Description:
Detailed description of function
Parameters: backend – Class instance Returns: model – Model with updated operators Note
Base class implements no modifications. This function is automatically called as a part of
validateModel.
-
-
Contents¶ DIAGONAL
- Files
- DiagonalAutoDiffBackend - Automatic differentiation backend class (diagonal representation) DiagonalJacobian - Diagonal representation of a Jacobian DiagonalSubset - Structured subset of a diagonal jacobian GenericAD - GenericAD is the testbed for future updates to the ADI class. All
-
class
DiagonalAutoDiffBackend(varargin)¶ Automatic differentiation backend class (diagonal representation)
Synopsis:
backend = DiagonalAutoDiffBackend()
Description:
This backend uses a diagonal representation (with an optional set of operators that produce intermediate diagonal representations). The primary use of this class is for problems with a large number of independent primary variables, with many cell-wise operations (e.g. compositional or similar problems).
Returns: Backend – Initialized class instance See also
AutoDiffBackend,SparseAutoDiffBackend-
modifyOperators= u'true'¶ Update the operators and use custom versions that return
DiagonalSubsetinstances
-
-
class
DiagonalJacobian(d, dim, subset, useMex)¶ Diagonal representation of a Jacobian
-
rdivide(u, v)¶ Right matrix divide:
h=u/v
-
diagonal= None¶ Dense matrix of diagonal derivatives
-
dim= None¶ Vector: First dimension is the number of variables in block, while the second is the number of columns
-
subset= None¶ Indices corresponding to the subset (if empty, class contains the full set)
-
-
class
DiagonalSubset(d, dims, map, subset, parentSubset)¶ Structured subset of a diagonal jacobian
-
map= None¶ Map to the underlying DiagonalJacobian representation. Two DiagonalSubsets of the same map can be multiplied together, etc.
-
-
class
GenericAD(val, jac, numVars, offset, useMex)¶ GenericAD is the testbed for future updates to the ADI class. All features herein are subject to rapid change.
-
interpPVT(T, x, v, flag)¶ Interpolate special PVT table with region support
-
max(u, v)¶ this function should be expanded
-
mtimes(u, v)¶ ‘*’
-
power(u, v)¶ ‘.^’
-
times(u, v)¶ ‘.*’
-
-
Contents¶ OPERATORS
- Files
- discreteDivergence - Discrete divergence for the GenericAD library faceAverage - Face average operator for the GenericAD library singlePointUpwind - Single-point upwind for the GenericAD library twoPointGradient - Discrete gradient for the GenericAD library
-
discreteDivergence(acc, N, v, nc, nf, sortIx, C, prelim, useMex)¶ Discrete divergence for the GenericAD library
-
faceAverage(N, v, useMex)¶ Face average operator for the GenericAD library
-
singlePointUpwind(flag, N, v, useMex)¶ Single-point upwind for the GenericAD library
-
twoPointGradient(N, v, M, useMex)¶ Discrete gradient for the GenericAD library
-
Contents¶ UTILS
- Files
- diagMult - Internal function for diagonal multiplication in AD code diagProductMult - Undocumented Utility Function double2GenericAD - Convert a double to GenericAD variable, using a sample GenericAD variable for dimensions getSparseArguments - Get sparse matrix indices getSparseBlocks - Get sparse blocks incrementSubset - Update a subset directly initVariablesAD_diagonal - Diagonal AD initializer initVariablesAD_oneBlock - Initialize a set of automatic differentiation variables (single block) matrixDims - Overloadable version of size
-
diagMult(v, M, D)¶ Internal function for diagonal multiplication in AD code
-
diagProductMult(v1, v2, x, y, D1, D2)¶ Undocumented Utility Function
-
double2GenericAD(u, sample)¶ Convert a double to GenericAD variable, using a sample GenericAD variable for dimensions
Synopsis:
u = double2GenericAD(u, adivar)
Parameters: - u – Double to be converted to GenericAD.
- sample – Sample variable of the type GenericAD to be used.
Returns: u – Variable with same type as sample and same value as u initially had. If u is a GenericAD class instance, u will have zero jacobians with the same number of primary variables as the jacobians of sample.
See also
GenericAD
-
getSparseArguments(M, ioffset, joffset)¶ Get sparse matrix indices
-
getSparseBlocks(varargin)¶ Get sparse blocks
-
incrementSubset(x, subs, v)¶ Update a subset directly
-
initVariablesAD_diagonal(varargin)¶ Diagonal AD initializer
-
initVariablesAD_oneBlock(varargin)¶ Initialize a set of automatic differentiation variables (single block)
-
matrixDims(varargin)¶ Overloadable version of size
-
Contents¶ SPARSE
- Files
- SparseAutoDiffBackend - Automatic differentiation backend class (sparse representation)
-
class
SparseAutoDiffBackend(varargin)¶ Automatic differentiation backend class (sparse representation)
Synopsis:
backend = SparseAutoDiffBackend()
Description:
This version of the AD backend uses different types of sparse blocks to represent derivatives.
Returns: Backend – Initialized class instance See also
AutoDiffBackend,DiagonalAutoDiffBackend-
updateDiscreteOperators(backend, model)¶ Do nothing
-
useBlocks= None¶ Organize Jacobian as a set of blocks, instead of one large AD matrix
-
Utilities¶
-
Contents¶ UTILS
- Files
- addPropertyDependence - Document dependencies and external dependencies addFluxesFromSourcesAndBC - Add in fluxes imposed by sources and face boundary conditions assignValue - Assign values to ADI object by way of indices, without changing jacobians bc2ADbc - INTERNAL DEPRECATED FUNCTION: Intentionally undocumented. checkWellConvergence - Compute convergence for wells. CNV_MBConvergence - Compute convergence based on total mass balance and maximum residual mass balance. combineEquations - Combine equations. For doubles, this is equivialent to a vertical compressSchedule - Compress schedule to take the longest possible timesteps while honoring controls computeCpGeometry - Undocumented Utility Function computeSourcesAndBoundaryConditionsAD - Compute phase-pseudocomponent source terms (compatible with AD codes) convert2MSWell - Utility for Converting Standard Well Structure to Multi-Segment Type convertDeckScheduleToMRST - Convert deck-type schedule to MRST style schedule convertIncompWellSols - Convert wellSols from incomp module to format used in ad-core/ad-blackoil convertReportToSchedule - Create a new schedule based on actual ministeps from a simulation report convertReservoirFluxesToSurface - Compute surface fluxes from reservoir fluxes crossFlowMixture - Undocumented Utility Function crossFlowMixtureDensity - Undocumented Utility Function double2ADI - Convert a double to ADI variable, using a sample ADI variable for dimensions estimateCompositionCFL - Undocumented Utility Function estimateSaturationCFL - Undocumented Utility Function expandIfUniform - Utility which reverses “value” compaction. If given a matrix (logical faceUpstr - Perform single-point upwinding of cell values to face fastInterpTable - Fast interpolation of table, using griddedInterpolant getBoundaryConditionFluxesAD - Get boundary condition fluxes for a given set of values getCellMajorReordering - Get equation ordering transforming variable major to cell major ordering getConvergenceValuesCNV - Compute convergence based on total mass balance and maximum residual mass balance. getConvergenceValuesWells - Undocumented Utility Function getFaceTransmissibility - Compute face transmissibilities, accounting for input-specific multipliers getFractionalFlowMagnitude - Undocumented Utility Function getGridSYMRCMOrdering - Undocumented Utility Function getMultiDimInterpolator - Get a multidimensional interpolator (with support for ADI varibles) getMultipliers - Get dynamic multiplier values for reservoir quantities getPerforationToWellMapping - Get map from global perforation number to global well index. getReportMinisteps - Get the timesteps used for the ministeps of a report getSampleAD - Utility for getting a AD value if it exists from a list of possible getSimulationTime - Get the global time for a set of states produced by simulateScheduleAD getSourceFluxesAD - Short description getWellOutput - Extract values from wellsols. HandleStruct - initWellSolAD - Set up well solution struct for a automatic differentiation model interpolateIDW - Undocumented Utility Function makeScheduleConsistent - Ensure that a schedule is consistent in terms of well counts/perforations mergeOrderedArrays - Merge two sets of cells that are similar in that they may contain numelData - Alias for numel. Useful for writing code which handles either numelValue - Undocumented Utility Function padRatesAndCompi - Pad one/two/threephase values with zeros corresponding to missing phases. phaseDensitiesTobfactor - Convert densities to b-facctors, accounting for dissolution pressureBCContrib - LEGACY FUNCTION: Intentionally undocumented. pressureBCContribADI - LEGACY FUNCTION: Intentionally undocumented. printConvergenceReport - Print a neatly formatted convergence report readSummaryLocal - Undocumented Utility Function recoverVars - Recover previously eliminated variables x at position n using solutions sol refineSchedule - Compute a finer schedule, including new time steps but preserving the time steps of the original reorderForILU - Attempt to reorder a set of equations so that the diagonal is non-zero ResultHandler - Class for storing and retrieving simulation results, either in memory or stored to disk selectLinearSolverAD - Undocumented Utility Function selectModelFromDeck - Select simulation model from a ECLIPSE/FrontSim style input deck setupOperatorsTPFA - Set up helper structure for solvers based on automatic differentiation. setWellSign - Ensure that wells have a defined sign. Will attempt to guess based on controls. simpleSchedule - Make a schedule with varying timesteps and fixed wells/bc/src terms splitFaceCellValue - Split multi-valued function into cell and face values splitMatrixForReduction - Split matrix A and right-hand side into blocks standaloneSolveAD - Solve a single time-step with AD solvers for given forces structPropEvaluated - Undocumented Utility Function terniaryWellPlot - Plot well curves (water, gas, oil and optionally BHP) for wellSols wellSolToVector - Extract selected summary vectors from cell array of well solutions
-
class
ResultHandler(varargin)¶ Class for storing and retrieving simulation results, either in memory or stored to disk
Synopsis:
handler = ResultHandler() handler = ResultHandler('dataPrefix', 'mydata', 'writeToDisk', true);
Description:
This class can be used to store and retrieve simulation results. It is somewhat similar to a cell array in use, although more limited.
Take a look at the class declaration to get more information.
Example:
% Setup handler handler = ResultHandler('dataprefix', 'mydata', 'writeToDisk', true); % Write result handler{1} = {'hello'}; % Read result from disk and print disp(handler{1})
Returns: Class instance that in some limited aspects acts like a cell array -
writeToFile(handler, data, id)¶ #ok
-
cleardir= None¶ Internal data storage
-
data= None¶ Flag indicating if verbose output is on. Will output storage and
-
dataDirectory= None¶ The folder under directory where we will store results. Will be
-
dataFolder= None¶ Data will be stored in the format <dataPrefix><index> so that
-
dataPrefix= None¶ Flags passed on to MATLAB builtin ‘save’. Consider ‘-v7’ if
-
saveflags= None¶ Clear directory on startup
-
storeInMemory= None¶ Directory where data in general is stored.
-
writeToDisk= None¶ Boolean indicating if the class should store results in memory
-
-
CNV_MBConvergence(model, problem)¶ Compute convergence based on total mass balance and maximum residual mass balance.
Synopsis:
[converged, values, evaluated] = CNV_MBConvergence(model, problem)
Description:
Compute CNV/MB type convergence similar to what is used for black oil convergence in commercial simulators.
Parameters: - model – Subclass of PhysicalModel. Strongly suggested to be some black oil variant, as this convergence function does not account for general residual convergence.
- problem – LinearizedProblem class instance we want to test for convergence.
Returns: - convergence – Boolean indicating if the state used to produce the LinearizedProblem has converged.
- values – 1 by 6 array containing mass balance in the first three terms followed by cnv in the last three. The phase ordering is assumed to be oil, water, gas. Phases present will return a zero in their place.
- evaluated – Logical array into problem.equations indicating which residual equations we have actually checked convergence for.
- names – Cell array of same length as values with short names for printing/debugging.
-
addFluxesFromSourcesAndBC(model, eqs, pressure, rho, mob, s, forces)¶ Add in fluxes imposed by sources and face boundary conditions
Description:
Utility function for updating residual conservation equations in the AD framework with additional fluxes due to boundary conditions and sources. Wells are handled separately in WellModel.
Parameters: - model – Simulation model (subclass of ReservoirModel).
- eqs –
- Residual conservation of mass-equations for each phase, in
- the order WATER, OIL, GAS (with any inactive phases omitted) as a cell array.
(All the following arguments are cell arrays, with length equal to the number of the active phases, with the values for each cell in each entry unless otherwise noted)
pressure - Phase pressures rho - Surface densities (one value per phase) mob - Phase mobilities s - Phase saturations- forces - Struct containing .src and .bc fields for sources and
- boundary conditions respectively.
Returns: - eqs – Phase conservation equations with added fluxes.
- qBC – Phase fluxes due to BC at standard conditions.
- BCTocellMap – Matrix mapping qBC to cells.
- qSRC – Phase fluxes due to source terms.
- srcCells – List of cells, mapping qSRC to cells.
See also
getBoundaryConditionFluxesAD, getSourceFluxesAD, addSource, addBC
-
addPropertyDependence(prop, name, grouping)¶ Document dependencies and external dependencies
-
assignValue(x, v, inx)¶ Assign values to ADI object by way of indices, without changing jacobians
Synopsis:
x = assignValue(x, v, inx)
Description:
Replace the numerical values of a ADI or double, without changing the Jacobians. This can lead to inconsistent Jacobians and variables so it should only be used if you really know what you are doing!
Parameters: - x – ADI or double where values are to be replaced.
- v – Values that will replace some subset of x.
- inx - The indices into x that v will replace. That is, after the call,
- double(x(ix)) == v
Returns: x – Modified version of input with same class. See also
ADI
-
bc2ADbc(G, bc)¶ INTERNAL DEPRECATED FUNCTION: Intentionally undocumented.
-
checkWellConvergence(model, problem)¶ Compute convergence for wells.
Synopsis:
[converged, values] = CNV_MBConvergence(model, problem)
Description:
- Compute convergence for well equations. Uses the properties
- model.toleranceWellRate moedl.toleranceWellBHP
Parameters: - model – Subclass of PhysicalModel that contain equations that are of type well and perforations.
- problem – LinearizedProblem class instance we want to test for convergence.
Returns: - convergence – Boolean indicating if the state used to produce the LinearizedProblem has converged.
- values – Residual inf of wells.
- evaluated – Logical array into problem.equations indicating which residual equations we have actually checked convergence for.
-
combineEquations(varargin)¶ Combine equations. For doubles, this is equivialent to a vertical concatenation. Please note that the full implementation used is found as a part of the ADI class. This is the fallback for doubles.
-
compressSchedule(schedule)¶ Compress schedule to take the longest possible timesteps while honoring controls
Synopsis:
schedule = compressSchedule(schedule)
Description:
Take a already defined schedule and combine all successive timesteps with the same controls. This is useful to make a schedule suitable for dynamic timestepping, as the control steps correspond only to the hard limits set by changing well/bc controls.
Parameters: scheduleDeck – A deck struct, typically from convertDeckScheduleToMRST or manually created. Returns: scheduleCompressed – Schedule ready for simulation in ‘simulateScheduleAD’.
-
computeCpGeometry(G, grdecl, varargin)¶ Undocumented Utility Function
-
computeSourcesAndBoundaryConditionsAD(model, pressure, s, mob, rho, dissolved, forces)¶ Compute phase-pseudocomponent source terms (compatible with AD codes)
Synopsis:
[src, bc] = computeSourcesAndBoundaryConditionsAD(model, pressure, s, mob, rho, dissolved, forces)
Parameters: - model – Subclass of ReservoirModel for which the source terms are to be computed.
- pressure – Reservoir pressures (cell array, one pressure per phase)
- s – Phase saturations (cell array, one saturation per phase)
- mob – Phase mobilities (cell array, one mobilit per phase)
- rho – Phase densities (including contributions from dissolved phases. For a black-oil style model, this is rhoO = bO.*(rs*rhoGS + rhoOS), and not the pseudocomponent density in the phase bO.*rhoOS!
- dissolved – Dissolution matrix for the properties. See
getDissolutionMatrixinThreePhaseBlackOilModel. - forces – Struct containing standard MRST driving forces.
Specifically, this routine uses the src and bc fields. All
other fields are ignored. For well source terms, see the
FacilityModel.
Returns: src,bc – Structs for sources and boundary conditions respectively. Each struct uses the same format with the following fields:
‘phaseMass’ - Cell array of mass source terms per phase pseudocomponent, accounting for dissolved fractions. ‘phaseVolume’ - Cell array of volumetric source terms per phase at reservoir conditions. ‘components’ - Empty cell array for inserting component source terms. Components are not the responsibility of this function, but we add the field to ensure that the structure is normalized. ‘mapping’ - Either empty or a matrix used to map the source terms into aggregate per-cell values. This matrix is required when multiple source terms are defined in the same block (e.g. two faces for a cell) since Matlab overwrites repeat indices instead of summing them. ‘sourceCells’ - List of cells the source terms should be added to.
Note
The practical implementation of boundary conditions is normally done through the gateway ReservoirModel>addBoundaryConditionsAndSources routine, which uses this routine directly.
See also
equationsOilWater,equationsBlackOil,ReservoirModel>addBoundaryConditionsAndSources
-
convert2MSWell(w, varargin)¶ Utility for Converting Standard Well Structure to Multi-Segment Type
Derives and includes Well Nodes and Well Segments in the well structure.
Synopsis:
W = convert2MSWell(W)
-
convertDeckScheduleToMRST(model, scheduleDeck, varargin)¶ Convert deck-type schedule to MRST style schedule
Synopsis:
schedule = convertDeckScheduleToMRST(model, deck.SCHEDULE)
Description:
Take a schedule in deck-style (from for example the output of readEclipseDeck), parse all wells and create a new schedule suitable for ‘simulateScheduleAD’.
Parameters: - G – Valid grid (likely from initEclipseGrid). Must be the same grid as the wells in the schedule are defined for.
- rock – Valid rock used to compute the well indices. Typically from initEclipseRock.
- scheduleDeck – Either a deck struct from readEclipseDeck or the schedule (typically deck.SCHEDULE).
Keyword Arguments: ‘StepLimit’ – Only parse the first n control steps.
Returns: scheduleMRST – Schedule ready for simulation in ‘simulateScheduleAD’.
-
convertIncompWellSols(W, states, incompFluid)¶ Convert wellSols from incomp module to format used in ad-core/ad-blackoil
Synopsis:
wellSols = convertIncompWellSols(W, states, fluid)
Description:
The solvers in the
incompmodule uses a different wellSol format than thead-corestyle wellSols. This function converts a set of states into wellSols suitable for routines that were designed to work with thead-corestyle wellSols. Specifically, this enables the use of getWellOutput and plotWellSols with solutions from the incompressible solvers not based on AD.Parameters: - W – Wells used for simulation
- states – Nstep long struct array of all simulation states.
- incompFluid – Fluid model used to compute the simulation states.
Returns: wellSols – Nstep long cell array of the same format as output by e.g.
simulateScheduleADSee also
-
convertReportToSchedule(report, schedule)¶ Create a new schedule based on actual ministeps from a simulation report
Synopsis:
schedule = convertReportToSchedule(report, schedule)
Description:
Running a simulation schedule with a given set of control steps may lead to several extra timesteps due to time step cutting/adjustments done by the nonlinear solver. This utility converts the report output from ‘simulateScheduleAD’ along with the schedule used into a new schedule that accounts for ministeps actually taken.
Note that ‘simulateScheduleAD’ MUST be called with the option ‘OutputMinisteps’ set to true for this to do anything, otherwise it will just output the report steps.
Parameters: - report – Report from ‘simulateScheduleAD’ with ‘OutputMinisteps’ set to true.
- schedule – The schedule used to produce the report.
Returns: schedule – New schedule modified so that the ministeps are accounted for.
See also
simulateScheduleAD
-
convertReservoirFluxesToSurface(model, states)¶ Compute surface fluxes from reservoir fluxes
Synopsis:
states = convertReservoirFluxesToSurface(model, states)
Description:
This function, given states with .bfactors and .flux, will compute the surface/standard condition fluxes and place them under the field surfaceFlux. To ensure bfactors and fluxes are added to states during a simulation, enable the flag “model.extraStateOutput” before simulating.
Parameters: - model – ReservoirModel subclass used to produce the states.
- states – States with valid fields .flux and .bfactors.
Returns: states – States with additional field surfaceFlux.
-
crossFlowMixture(flux, compi, map, conserveMass, is_zero)¶ Undocumented Utility Function
-
crossFlowMixtureDensity(massFlux, volumeTotalFlux, massFluxFromSurface, map)¶ Undocumented Utility Function
-
double2ADI(u, sample)¶ Convert a double to ADI variable, using a sample ADI variable for dimensions
Synopsis:
u = double2ADI(u, adivar)
Parameters: - u – Double to be converted to ADI.
- sample – Sample variable of the type ADI to be used.
Returns: u – Variable with same type as sample and same value as u initially had. If u is a ADI class instance, u will have zero jacobians with the same number of primary variables as the jacobians of sample.
See also
ADI, initVariablesADI
-
estimateCompositionCFL(model, state, dt, varargin)¶ Undocumented Utility Function
-
estimateSaturationCFL(model, state, dt, varargin)¶ Undocumented Utility Function
-
expandIfUniform(v)¶ Utility which reverses “value” compaction. If given a matrix (logical or numerical) as input, it will expand it to a cell array of vectors such that value(expandIfUniform(x)) is equal to x.
-
faceUpstr(flag, x, N, sz)¶ Perform single-point upwinding of cell values to face
Synopsis:
[xu, xc] = faceUpstr(flag, x, N, sz)
Description:
Perform single-point upwind. A robust discretization for transported/hyperbolic variables.
Parameters: - flag – Boolean for each face indicating if the face should take the value from the first cell (if true), or the second cell (if false).
- x – Vector of values to be upwinded. One value per cell in the
domain. See
szinput. - N – Neighborship. A number of faces by 2 array. Each row corresponds to the cells connected to the face with that number.
- sz – Vector of length 2. First entry corresponds to the number of faces and the second is the total number of cells.
-
fastInterpTable(X, Y, xi)¶ Fast interpolation of table, using griddedInterpolant
Synopsis:
yi = fastInterpTable(X, Y, xi)
Description:
A simple wrapper for griddedInterpolant for fast interpolation of simple data in the AD-framework. Always defaults to linear interpolation with linear extrapolation.
Parameters: - x – Sample X-coordinates
- y – Sample function values
- xi – The X-coordinates at which the linear interpolant is to be evaluated.
Returns: yi – Linear function interpolating (x, y) evaluated at xi.
-
getBoundaryConditionFluxesAD(model, pressure, s, mob, rho, b, bc)¶ Get boundary condition fluxes for a given set of values
Synopsis:
[qSurf, BCTocellMap, BCcells] = getBoundaryConditionFluxesAD(model, pressure, s, mob, rho, b, bc)
Description:
Given a set of boundary conditions, this function computes the fluxes induced for a given set of reservoir parameters (density, mobility, saturations etc).
Parameters: - model – Subclass of ReservoirModel implementing the current simulation model.
- pressure – Cell values of pressure. Should be a nph long cell array, containing the phase pressures.
- rho – Surface densities of each phase, as a nph long cell array.
- s – Phase saturations per cell, as a nph long array.
- bc – Boundary condition struct, with valid .sat field with length nph. Typically made using addBC, pside or fluxside.
Returns: - qSurf – Cell array of phase fluxes.
- BCTocellMap – Matrix used to add in bc fluxes to cells. Implemented as a matrix to efficiently account for cells with multiple faces with boundary conditions.
- cells – The cells affected by boundary conditions.
See also
addBC, pside, fluxside
-
getCellMajorReordering(ncell, block_size, varargin)¶ Get equation ordering transforming variable major to cell major ordering
Synopsis:
ordering = getCellMajorReordering(ncell, block_size)
Description:
Get a permutation vector which transforms a system on the standard variable major (e.g., a two-phase system p_1, …, p_n, s_1, …, s_n) into a cell major (e.g. p_1, s_1, …, p_n, s_n) where p and s are two primary variables and the subscript refers to a specific cell.
If Ax=b is some system to be re-ordered of size ncell*block_size, then ordering = getCellMajorReordering(ncell, block_size); A = A(ordering, ordering); b = b(ordering); will re-order the system. Solving the system x = solve(A, b) where solve is some linaer solver will then give a permuted solution to the system. The final solution is then x(ordering) = x.
The primary utility of this function is to a) Allow the user to call external linear solvers which require this type of ordering and b) change the system ordering for e.g. ILU(0).
Parameters: - ncell – Number of cells in grid to be used.
- block_size – Size of each block (e.g. number of components present)
Keyword Arguments: - ndof – Total number of degrees of freedom. Any additional degrees of freedom beyond the ncell*block_size first variables will have a identity remapping, retaining the position in the final system.
- equation_ordering – An optional ordering of the equations. Should be a block_size length vector.
- cell_ordering – An optional ordering of the cells themselves.
Returns: ordering – Ordering vector so that A = A(ordering, ordering) is the permuted system
See also
LinearSolverAD,AMGCL_CPRSolverAD
-
getConvergenceValuesCNV(model, problem)¶ Compute convergence based on total mass balance and maximum residual mass balance.
Synopsis:
[converged, values, evaluated] = CNV_MBConvergence(model, problem)
Description:
Compute CNV/MB type convergence similar to what is used for black oil convergence in commercial simulators.
Parameters: - model – Subclass of PhysicalModel. Strongly suggested to be some black oil variant, as this convergence function does not account for general residual convergence.
- problem – LinearizedProblem class instance we want to test for convergence.
Returns: - convergence – Boolean indicating if the state used to produce the LinearizedProblem has converged.
- values – 1 by 6 array containing mass balance in the first three terms followed by cnv in the last three. The phase ordering is assumed to be oil, water, gas. Phases present will return a zero in their place.
- evaluated – Logical array into problem.equations indicating which residual equations we have actually checked convergence for.
- names – Cell array of same length as values with short names for printing/debugging.
-
getConvergenceValuesWells(model, problem)¶ Undocumented Utility Function
-
getFaceTransmissibility(G, rock, deck, varargin)¶ Compute face transmissibilities, accounting for input-specific multipliers
Synopsis:
T = getFaceTransmissibility(G, rock, deck) T = getFaceTransmissibility(G, rock)
Description:
Computes transmissibilities per interface. Accounts for multipliers due to both general transmissibility multipliers and fault multipliers specifically.
Parameters: - G – Valid grid structure.
- rock – Valid rock structure.
- deck – (OPTIONAL) ECLIPSE style input deck used to produce the grid, typically produced by readEclipseDeck. Needed to account for multipliers.
Keyword Arguments: (Passed directly onto underlying function computeTrans)
Returns: T – Transmissibilities, one per interface.
See also
computeTrans
-
getFractionalFlowMagnitude(model, state)¶ Undocumented Utility Function
-
getGridSYMRCMOrdering(G)¶ Undocumented Utility Function
-
getMultiDimInterpolator(x, Fx, extrap)¶ Get a multidimensional interpolator (with support for ADI varibles)
Synopsis:
fn = getMultiDimInterpolator(x, Y);
Parameters: - x – Cell array containing the vectors for the points of the function. Must have equal length to the number of dimensions in Y.
- Y – A matrix of dimension equal to x, representing a structured dataset for multdimensional interpolation
Returns: - fn – A function for interpolation with variable arguments equal to the length of x.
- F – griddedInterpolant class instace
- epsilons – Epsilon values for each input argument representing one half of the minimum distance between elements. Useful for computing numerical derivatives of the interpolant since Matlab does not expose the slopes.
See also
-
getMultipliers(fluid, p, p0)¶ Get dynamic multiplier values for reservoir quantities
Synopsis:
[pvMult, transMult, mobMult] = getMultipliers(fluid, p, p0)
Parameters: - fluid – Fluid model, typically from initDeckADIFluid or some other constructor.
- p – Pressure per cell.
- p0 – Pressure per cell (previous timestep).
Returns: - pvMult – Pore volume multiplier per cell. Multiplicative modifier for
the pore volume based on a simplified model for how the pores grow when fluid pressure is increasing.
- transMult - Transmissibility multiplier, modelling pressure dependent
permeability. One value per interface.
mobMult - Mobility multiplier per cell.
pvMult0, transMult0, mobMult0 - Multipliers for previous timestep.
-
getPerforationToWellMapping(w)¶ Get map from global perforation number to global well index.
Synopsis:
perf2well = getPerforationToWellMapping(w)
Parameters: w – Well structure. Returns: perf2well – perf2well(ix) will give the well number of global perforation number ix. See also
WellModel
-
getReportMinisteps(report)¶ Get the timesteps used for the ministeps of a report
Synopsis:
timesteps = getReportMinisteps(report)
Description:
Get the actual ministeps used by simulateScheduleAD.
Parameters: report – Report from simulateScheduleAD. Returns: timesteps – The timesteps used to solve the problem. These differ from the control steps (schedule.step.val) in that they are the actual timesteps taken by the solver (due to timestep cutting, adaptive timestepping and so on) See also
SimulateScheduleAD
-
getSampleAD(varargin)¶ Utility for getting a AD value if it exists from a list of possible AD-values
-
getSimulationTime(states, report)¶ Get the global time for a set of states produced by simulateScheduleAD
Synopsis:
times = getSimulationTime(states, report)
Description:
Get the time for each state output by simulateScheduleAD.
Parameters: - states – Cell array of states as given by simulateScheduleAD. Can be either per control step or per ministep.
- report – Report given by simulateScheduleAD.
Returns: times – Array with same dimensions as states, giving the values of the time in the simulation model for each step. The initial state passed to simulateScheduleAD is assumed to be at time zero.
See also
simulateScheduleAD
-
getSourceFluxesAD(model, mob, s, src)¶ Short description
Synopsis:
[qSurf, BCTocellMap, cells] = getSourceFluxesAD(model, mob, b, s, src)
DESCRIPTION:
Parameters: - model – Subclass of ReservoirModel indicating which phases are active.
- mob – A cell array of cell mobility values for all active phases.
- s – A cell array of saturations per cell for all active phases.
- src – Source struct as defined by addSource
Returns: - qSurf – Source terms at standard conditions. Cell array of same dimensions as the number of active phases.
- cells – A list of cells for which the entries of qRes should be added.
-
getWellOutput(wellsols, fldnames, wells)¶ Extract values from wellsols.
Synopsis:
[wd, wn, flds]= getWellOutput(wellSols, 'bhp') [wd, wn, flds]= getWellOutput(wellSols, {'bhp', 'qWs'}, 'W1')
Description:
Given a cell array of well solution structures representing multiple timesteps, this routine extracts requested values in matrix form ready for plotting / inspection.
Parameters: - wellSols – Cell array of NSTEP length, each containing a struct with NWELL entries. All wells must exist at all timesteps.
- fldnames – (OPTIONAL) Either a single string, or a cell array of desired fields for output. Bottom hole pressures, rates, … Defaults to all fields.
- wells – (OPTIONAL) Either a single string, or a cell array of well names for which output is desired. Defaults to all wells.
Returns: - welldata – A NSTEP by NWELL by NFIELDS matrix. For instance, for
- calling
- D = getWellOutput (wellsols, {‘bhp’, ‘qWs’}, {‘Injector’, ‘Producer’})
- will give bottom hole pressures for the producer in D( – , 1, 2);
See also
plotWellSols
-
initWellSolAD(W, model, state0, wellSolInit)¶ Set up well solution struct for a automatic differentiation model
Synopsis:
wellSol = initWellSolAD(W, model, state0); wellSol = initWellSolAD(W, model, state0, ws);
Description:
Create or extract the wellSol, and ensure that it contains the correct fields for advanced solvers with well limits and variable perforation counts. This function will first look for a explicitly passed wellSol to modify, then it will consider any wellSol residing in state0. If neither is found, it will attempt to construct one based on W.
Parameters: - W – Control well for which we are going to create a well solution structure.
- model – Subclass of ReservoirModel. Used to determine how many and which phases are present.
- state0 – State, possibly with a wellSol given already (see initResSol/initState).
- wellSolInit – Initial wellSol.
Returns: wellSol – Well solution struct with additional fields ready for simulation.
-
interpolateIDW(x, f, xq, order)¶ Undocumented Utility Function
-
makeScheduleConsistent(schedule, varargin)¶ Ensure that a schedule is consistent in terms of well counts/perforations
Synopsis:
schedule = makeScheduleConsistent(schedule)
Description:
For a given schedule with varying amount of wells and perforated cells per well, this schedule makes the schedule internally consistent so that all wells are defined at each control step. Some wells will be disabled at different points, but they are always present and thus the simulator output will be normalized and easier to work with.
Parameters: schedule – Schedule with possibly inconsistent numbers of wells and perforations. Returns: schedule – Equivialent schedule that is consistent in the well and cell numberings. See also
convertDeckScheduleToMRST
-
mergeOrderedArrays(old, new)¶ Merge two sets of cells that are similar in that they may contain elements from the same superset in the same order, but each set may be missing one or more elements that the other has.
This is done by having two simple pointers that are incremented as we go along trying to merge the two sets.
-
numelData(x)¶ Alias for numel. Useful for writing code which handles either ResultHandler or cell arrays.
-
numelValue(v)¶ Undocumented Utility Function
-
padRatesAndCompi(q_s, W, model)¶ Pad one/two/threephase values with zeros corresponding to missing phases.
Synopsis:
[q, W] = padRatesAndCompi(q, W, model);
Description:
This function adds padding zeros to convert rates and wells for a one/two phase model to make it appear as a three phase model with zero rates for the missing phases.
Parameters: - q_s – Cell array of fluxes corresponding to the number of active phases in the model.
- W – Wells compatible with the current model.
- model – Model with one or more active phases, consistent with q_s and W.
Returns: - q_s – 1 by 3 cell array with zero values added for missing phases.
- W – Three phase wells (.compi contains three fields, again with zeros where phases are missing in the original model).
- isActive – Indicators for which phases are present.
See also
PhysicalModel, WellModel
-
phaseDensitiesTobfactor(rho, rhoS, dissolved)¶ Convert densities to b-facctors, accounting for dissolution
-
pressureBCContrib(G, s, pX, rhoX, mobX, bX, bc)¶ LEGACY FUNCTION: Intentionally undocumented.
-
pressureBCContribADI(G, s, pX, rhoX, mobX, bX, bc)¶ LEGACY FUNCTION: Intentionally undocumented.
-
printConvergenceReport(names, values, converged, iteration, endOfBlock)¶ Print a neatly formatted convergence report
Synopsis:
printConvergenceReport({'myEquation', 'yourEquation'}, [1, 25], [true, false], it);
Description:
Print convergence report to the Command Window. Two lines are plotted for the first iteration, and one line for succeeding iterations.
Parameters: - names – Names of the different convergence measures. Cell array of length N where N is the number of different measures (for instance, residual norms for different equations)
- values – Double array of length N, where each entry corresponds to the current value of the different named measures.
- converged – Boolean for each value indicating if convergence has been achieved for that value.
Returns: Nothing.
-
readSummaryLocal(prefix, keyWords)¶ Undocumented Utility Function
-
recoverVars(eq, n, sol)¶ Recover previously eliminated variables x at position n using solutions sol
-
refineSchedule(init_time_for_new_time_steps, new_time_steps, schedule)¶ Compute a finer schedule, including new time steps but preserving the time steps of the original schedule
Synopsis:
new_schedule = refineSchedule(init_time_for_new_time_steps, new_time_steps, schedule)
Parameters: - init_time_for_new_time_steps – Time where the sequence of new time steps will be added.
- new_time_steps – Sequence of time steps to be added.
- schedule – Input schedule which will be refined
-
reorderForILU(A, b, nc)¶ Attempt to reorder a set of equations so that the diagonal is non-zero
Synopsis:
[A, b] = reorderForILU(A, b, nc);
Description:
Reorder a set of equations to ensure non-zero diagonal. This is useful when building ILU-based solvers. Notably, this utility is useful whenever well equations are added, that may not have derivatives with respect to all well controls.
Parameters: - A – Linear system to be reordered.
- b – Right hand side of the system
- nc – (OPTIONAL) The routine will only look at equations from nc+1 and onwards to numel(b).
Returns: - A – Reordered linear system
- b – Reordered right hand side
-
selectLinearSolverAD(model, varargin)¶ Undocumented Utility Function
-
selectModelFromDeck(G, rock, fluid, deck, varargin)¶ Select simulation model from a ECLIPSE/FrontSim style input deck
Synopsis:
model = selectModelFromDeck(G, rock, fluid, deck)
Description:
Determine the type of PhysicalModel subclass (if any) most suitable for simulating a given input deck.
Parameters: - G – Simulation grid, typically from initEclipseGrid
- rock – Corresponding rock structure, typically from initEclipseRock.
- fluid – Fluid model, typically from initDeckADIFluid.
- deck – Parsed input deck, typically from readEclipseDeck.
Keyword Arguments: Any – Any extra arguments passed onto the model constructor directly.
Returns: model – Subclass of
PhysicalModelapproprioate for passing along tosimulateScheduleAD.See also
ThreePhaseBlackOilModel,TwoPhaseOilWaterModel,OilWaterPolymerModel
-
setWellSign(W)¶ Ensure that wells have a defined sign. Will attempt to guess based on controls.
Synopsis:
W = setWellSign(W)
Description:
The AD based solvers assume a W.sign is defined. This routine attempts to ensure that wells do have a sign. A positive sign is used to indicate an injector and a negative sign for a producer. If the wells have rate controls, they will be given signs based on the signs of the rates. If they are pressure controlled wells, they will get sign 0.
Parameters: W – Well struct, from e.g. addWell. Returns: W – Well struct where numel(W(i).sign) is guaranteed to be 1.
-
setupOperatorsTPFA(G, rock, varargin)¶ Set up helper structure for solvers based on automatic differentiation.
Synopsis:
s = setupOperatorsTPFA(G, rock)
Description:
The automatic differentiation solvers rely on discrete operators for divergence and gradient on the grid as well as a variety of derived reservoir quantities such as transmissibility and pore volume. The purpose of this function is to assemble all such quantities using a standard two-point finite volume approxiation (TPFA).
Parameters: - G – MRST grid given as a struct. See grid_structure.m for more details.
- rock – Rock structure containing fields .perm and .poro with approprioate dimensions for the grid. See makeRock for more details.
Keyword Arguments: - ‘deck’ – deck file containing rock properties
- ‘trans’ – transmissibility for internal faces (if neighbors given) or for all faces (if neighbors are not given)
- ‘neighbors’ – neighbors for each internal face
- ‘porv’ – pore volumes for all cells
Returns: - s – Operators struct, with discrete operators and derived
quantities:
T_all - Transmissibilities for all interfaces, including
(half) transmissibilities for faces on the boundary. One
value per interface.
T - Transmissibilities for all internal interfaces. Internal
interfaces have a cell on both sides.
pv - Pore volumes. See function
poreVolume. One value per cell. C - Transfer matrix between cells and faces. Used to derive discrete gradient and divergence operators. faceAvg - (Function) For each interface, computes the average value of a quantity defined in the cells. If a face is connecting two cells, the faceAvg function will compute the arithmetic average of the values in both cells. - Grad – Discrete gradient as function handle. Computes the gradient on each interface via a first order finite difference approximation using the values of the cells connected to the face. Note that this discrete gradient does not divide by the distance between the points.
- Div – (Function) Discrete divergence. Integrates / sums up values on the interfaces for all cells to give the (integrated) divergence per cell.
- faceUpstr – (Function) Perform upstream weighting of values. Given a set of cell wise values and a upstream flag for each interface, this function will pick the values corresponding to the position in the neighborship. I.e. if the flag is true for a given interface, it will select the value in the FIRST cell connected to the interface x(N(faceNo, 1)). Otherwise, it will select the SECOND x(N(faceNo, 2)). Typical usage is for upstream weighting of transported quantities.
- N – Neighborship structure. Will be number of interfaces by 2 in size where N(ix, :) contains the cells connected to face number ix.
-
simpleSchedule(dt, varargin)¶ Make a schedule with varying timesteps and fixed wells/bc/src terms
Synopsis:
schedule = simpleSchedule(timesteps); schedule = simpleSchedule(timesteps, 'W', W, 'src', src, 'bc', bc);
Parameters: dt – Vector (column/row) of desired timesteps.
Keyword Arguments: - W – Wells to be used in the schedule. The wells will be active in all timesteps.
- BC – Boundary conditions to be used in the schedule. The boundary conditions will be active in all timesteps.
- src – Source terms to be used in the schedule. The sourceterms will be active in all timesteps.
Returns: schedule – struct suitable for further modification, or for input to
simulateScheduleAD.See also
-
splitFaceCellValue(operators, flag, x, sz)¶ Split multi-valued function into cell and face values
Synopsis:
[fx, cx] = splitFaceCellValue(operators, flag, x, sz)
Description:
Taking a set of values, this function returns cell and face-upwinded values based on specified flag and dimensions. Normally, this is simply applying a pre-existing upwind operator to get the upstream weighted values for transported quantities. For special functions that arise in some workflows, it can take e.g. a set of (half)face values plus cell values and divide them up in a reasonable manner.
Parameters: - operators – Operators struct. See
setupOperatorsTPFA. - flag – Upstream flag to be used to upwind values. See
faceUpstr. - x – Vector of values to be treated. Can be either one value per cell in the domain, one value per face followed by one value per cell, or one value per half-face, followed by the cell values.
- sz – Vector of length 2. First entry corresponds to the number of faces and the second is the total number of cells.
See also
- operators – Operators struct. See
-
splitMatrixForReduction(A, b, n, strategy, doFactor)¶ Split matrix A and right-hand side into blocks A = [B, C] b = [f]
[D, E] [h]
-
standaloneSolveAD(state0, model, dt, varargin)¶ Solve a single time-step with AD solvers for given forces
Synopsis:
[state, report] = standaloneSolveAD(state0, model, dt, 'W', W, 'maxIterations', 25)
Description:
Stand-alone solver for AD. Useful for simple problems where a full schedule is not required. Calls simulateScheduleAD internally.
Parameters: - state0 – Initial state.
- model – PhysicalModel instance for simulation.
- dt – Timestep length.
Keyword Arguments: ‘Various’ – Additional inputs are given to the following functions in order of priority: First, as possible driving forces, then as inputs to simulateScheduleAD.
Returns: - state – Updated state.
- report – Reports for simulator.
See also
-
structPropEvaluated(s, name)¶ Undocumented Utility Function
-
terniaryWellPlot(wellSols, T, ix, varargin)¶ Plot well curves (water, gas, oil and optionally BHP) for wellSols
Synopsis:
% Plot well #3, with timesteps on xaxis terniaryWellPlot(wellSols, time, 3); % Plot all wells terniaryWellPlot(wellSols)
Description:
This function is tailored towards three-phase simulation and is capable of producing plots that include production rates for each well for each phase (water, oil gas) and optionally also bottom hole pressures as a separate axis. One figure is produced per well requested.
Parameters: - wellSols – Cell array of NSTEP by 1, each containing a uniform struct array of well solution structures. For example, the first output from simulateScheduleAD. Can also be a cell array of such cell arrays, for comparing multiple simulation scenarios.
- time – (OPTIONAL) The time for each timestep. If not provided, the plotter will use step number as the x axis intead. If wellSols is a cell array of multiple datasets, time should also be a cell array, provided not all datasets use the same timesteps.
- ix – (OPTIONAL) A list of indices to plot, or a single string corresponding to the name of a specific well. The default is all wells.
Keyword Arguments: ‘plotBHP’ – Boolean indicating if BHP is to be plotted. Defaults to enabled.
Returns: fh – Figure handles to all figures that were created.
See also
-
wellSolToVector(wellsols)¶ Extract selected summary vectors from cell array of well solutions
Synopsis:
[WaterRate, OilRate, GasRate, BHP] = wellSolToVector(wellSols)
Parameters: wellSols – Cell array of well solution structures as produced by runScheduleADI or simulateScheduleADI. Each solution structure must define the fields ‘qWs’, ‘qOs’, ‘qGs’, and ‘bhp’. Note
Function wellSolToVector does not support variable number of wells.
Returns: - In the following ‘nw’ refers to the number of wells
- (NUMEL (wellSols{1}))
- steps (NUMEL(wellSols))
- WaterRate – Numeric array of size nt-by-nw of water rate at surface conditions (unit m^3/s).
- OilRate – Numeric array of size nt-by-nw of oil rate at surface conditions (unit m^3/s).
- GasRate – Numeric array of size nt-by-nw of gas rate at surface conditions (unit m^3/s).
- BHP – Numeric array of size nt-by-nw of well bottom-hole pressure values (unit Pascal).
See also
simulateScheduleADI, runScheduleADI.
Examples¶
Classic Buckley-Leverett Problem: 1D Horizontal Displacement¶
Generated from adBuckleyLeverett1D.m
This example uses the classical Buckley-Leverett problem to introduce you to basic functionality in the object-oriented automatic differentiation framework. The Buckley-Leverett problem describes an incompressible displacement in a 1D homogeneous and horizontal medium with water injected at the left boundary x=0 and fluids produced at the right boundary x=L. We assume that the fluids have unit viscosity and density and have relative permeabilities that follow a standard quadratic Corey model
mrstModule add ad-blackoil ad-core ad-props mrst-gui
Set up model¶
We start by generating a model object that describes the reservoir. To construct this model, we need three more fundamental structures: ‘G’ represents the grid with reservoir geometry, ‘rock’ holds the petrophysical properties, and ‘fluid’ gives the fluid properties. In addition to the model object, we need a ‘state’ struct that defines the initial state in the reservoir (pressure and fluid saturations and compositions).
% Construct 3D grid with 50 cells in the x-direction
G = cartGrid([50, 1, 1], [1000, 10, 10]*meter);
G = computeGeometry(G);
% Homogenous rock properties
rock = makeRock(G, 1*darcy, .3);
% Default oil-water fluid with unit values
fluid = initSimpleADIFluid('phases', 'WO', 'n', [2 2]);
% Set up model and initial state.
model = TwoPhaseOilWaterModel(G, rock, fluid);
state0 = initResSol(G, 50*barsa, [0, 1]);
% Set up drive mechanism: constant rate at x=0, constant pressure at x=L
pv = poreVolume(G, rock);
injRate = sum(pv)/(500*day);
bc = fluxside([], G, 'xmin', injRate, 'sat', [1, 0]);
bc = pside(bc, G, 'xmax', 0*barsa, 'sat', [0, 1]);
Processing Cells 1 : 50 of 50 ... done (0.00 [s], 1.17e+04 cells/second)
Total 3D Geometry Processing Time = 0.004 [s]
Simulate 1 PVI using a manual loop¶
There are several ways to run a simulation. A simple approach is to use a manual loop, where you explicitly call a nonlinear solver to solve each implicit time step
solver = NonLinearSolver();
% Validate the model to prepare for simulation
model = model.validateModel();
% Validate the initial state
state0 = model.validateState(state0);
n = 25;
dT = (500/n)*day;
states = cell(n+1, 1);
states{1} = state0;
solver.verbose = true;
for i = 1:n
state = solver.solveTimestep(states{i}, dT, model, 'bc', bc);
states{i+1} = state;
end
Missing field "rs" added since disgas is not enabled.
Missing field "rv" added since vapoil is not enabled.
====================================================
| It # | CNV_W | CNV_O | MB_W | MB_O |
====================================================
| 1 | 2.00e+00 | 1.42e-01 |*2.31e-08 |*1.64e-09 |
| 2 | 1.72e+00 | 1.08e+00 |*2.08e-08 |*2.08e-08 |
| 3 | 6.40e-01 | 1.76e+00 |*1.62e-08 |*1.62e-08 |
...
Plot the result¶
We set up a plot using plotToolbar from the mrst-gui module. Since the problem is essentially one dimensional, we plot the water saturation (first column of the “s” field in state) as a 1D plot.
close all
plotToolbar(G, states, 'field', 's:1', 'plot1d', true, ...
'lockCaxis', true, 'startplayback', true);
Repeat simulation with general solver¶
To use the general solver, we first need to set up a schedule that describes the time steps and the drive mechanisms (wells, boundary conditions, and source terms) that are active in each time step. In addition, one can specify various forms of time-step control. Here, however, we simply rely on the default setup
schedule = simpleSchedule(repmat(dT,1,n), 'bc', bc);
[~,sstates] = simulateScheduleAD(state0, model, schedule);
close all
plotToolbar(G, sstates, 'field', 's:1','lockCaxis',true),
caxis([0 1]), view(10,10)
colorbar
*****************************************************************
********** Starting simulation: 25 steps, 500 days *********
*****************************************************************
Preparing model for simulation...
Model ready for simulation...
Preparing schedule for simulation...
Schedule ready for simulation...
Validating initial state...
...
Repeat simulation with visualization¶
The general solver has a hook, that enables you to visualize the progress of the simulation (and stop it and continue running it in ‘debug’ mode).
close all
fn = getPlotAfterStep(state0, model, schedule, ...
'plotWell', false, 'plotReservoir', true, 'field', 's:1', ...
'lockCaxis',true, 'plot1d', true);
[~,sstates,report] = ...
simulateScheduleAD(state0, model, schedule,'afterStepFn', fn);
*****************************************************************
********** Starting simulation: 25 steps, 500 days *********
*****************************************************************
Preparing model for simulation...
Model ready for simulation...
Preparing schedule for simulation...
Schedule ready for simulation...
Validating initial state...
...
Repeat simulation with a storage handler¶
The solver also has a hook that enables you to control how the computed states are stored. To control the storage, we use a so-called storage handler, which behaves almost like a cell array, whose content can either be stored in memory, on disk, or both places. Here, we set it up to store on disk
handler = ResultHandler('writeToDisk', true, 'dataFolder', 'AD-BL1D');
simulateScheduleAD(state0, model, schedule, 'outputHandler', handler);
disp(handler)
*****************************************************************
********** Starting simulation: 25 steps, 500 days *********
*****************************************************************
Preparing model for simulation...
Model ready for simulation...
Preparing schedule for simulation...
Schedule ready for simulation...
Validating initial state...
...
The simulation above stored each state as a separate *.mat file in a subdirectory ‘AD-BL1D’ somewhere on a standard path relative to MRST’s root directory. The time steps can now be accessed in the usual way as if they were in memory. We can, for instance, pass the handler to the visualization GUI:
figure
plotToolbar(G, handler, 'field', 's:1','lockCaxis', true, 'plot1d', true);
At a later stage, we can go back and retrieve the data from disk by constructing a new handler pointing to the same folder
clear handler
handler = ResultHandler('dataFolder', 'AD-BL1D');
m = handler.numelData();
states = cell(m, 1);
for i = 1:2:m
states{i} = handler{i};
end
If we do not want to keep the data, they can be removed by a call to the reset function:
handler.resetData()
Copyright notice¶
<html>
% <p><font size="-1
Managing simulations: Restart, packed problems and more¶
Generated from demoPackedProblems.m
When using MRST to simulate larger or many problems, it is best to ensure that the results are stored to disk in a predictible manner. Otherwise, output from a long simulation may be lost if Matlab is closed for whatever reason.
mrstModule add ad-core ad-blackoil ad-props mrst-gui
Set up a simple problem with quadratic and linear relperm¶
We set up a simple 1D displacement problem for purposes of this demonstration. We create the same scenario with two different fluid models: A version with linear relative permeability and one with quadratic.
name = '1d';
G = cartGrid([100, 1], [1000, 1000]);
rock = makeRock(G, 0.1*darcy, 0.3);
G = computeGeometry(G);
fluid_1 = initSimpleADIFluid('mu', [1, 1]*centi*poise, 'n', [1, 1], 'phases', 'wo', 'rho', [1000, 500]);
model_1 = TwoPhaseOilWaterModel(G, rock, fluid_1);
time = 10*year;
irate = sum(model_1.operators.pv)/time;
W = [];
W = addWell(W, G, rock, 1, 'type', 'rate', 'val', irate, 'comp_i', [1, 0]);
W = addWell(W, G, rock, G.cells.num, 'type', 'bhp', 'val', 100*barsa, 'comp_i', [1, 0]);
dt = rampupTimesteps(time, time/100);
schedule = simpleSchedule(dt, 'W', W);
state0 = initResSol(G, 150*barsa, [0, 1]);
% Second fluid model
fluid_2 = initSimpleADIFluid('mu', [1, 1]*centi*poise, 'n', [2, 2], 'phases', 'wo', 'rho', [1000, 500]);
model_2 = TwoPhaseOilWaterModel(G, rock, fluid_2);
Define two simulations¶
We create two atomic packed simulation problems. This is similar to using simulateScheduleAD, with the addition of a few parameters describing the problem.
% Define base name for all problems in this script
BaseName = 'test_packed';
% Define linear flux
problem_1 = packSimulationProblem(state0, model_1, schedule, BaseName, ...
'Name', 'linear_relperm', ...
'Description', '1D displacement with linear flux');
% Define nonlinaer flux
problem_2 = packSimulationProblem(state0, model_2, schedule, BaseName, ...
'Name', 'quadratic_relperm', ...
'Description', '1D displacement with non-linear flux');
Run simulation¶
This will only simulate what is not yet simulated, and can restart aborted simulations in a seamless manner by using the ResultHandler class internally.
problems = {problem_1, problem_2};
[ok, status] = simulatePackedProblem(problems);
We can get output, just as if we simulated in the standard manner¶
[ws_1, states_1, reports_1] = getPackedSimulatorOutput(problem_1);
[ws_2, states_2, reports_2] = getPackedSimulatorOutput(problem_2);
For many packed problems, we can also get the data in a single call¶
[all_ws, all_states, all_reports, names, report_time] = getMultiplePackedSimulatorOutputs(problems);
Plot well results¶
plotWellSols(all_ws, report_time, 'DatasetNames', names, 'field', 'qWs', 'SelectedWells', 2);
Remove data (with prompts)¶
clearPackedSimulatorOutput(problems)
We can also launch simulations in the background¶
This is useful for running multiple cases. This functionality does not require any Matlab add-ons, since it creates an additional Matlab session which runs in the background. Please note that the additional sessions will terminate upon completion on error, but for larger cases this may take some time. You should not launch more sessions than your workstation can handle! First, remove data (without prompt before deletion)
learPackedSimulatorOutput(problems, 'prompt', false)
% Launch simulations
for i = 1:numel(problems)
info = simulatePackedProblemBackground(problems{i}, 'verbose', true);
end
% Monitor the simulations running in the background
monitorBackgroundSimulations(problems);
% <html>
% <p><font size="-1
The use of regions: Different functions in different parts of the domain¶
Generated from differentRegionFunctionsExample.m
In many applications there is a need for varying functions in different parts of the domain. A typical example of this is the problem of varying rock types, where multiphase flow happens in a medium where the surface properties and porous structure varies significantly in different regions of the domain. In this example we demonstrate how to set up multiple rock types for relative permeability. The purpose of this example is to show the function interfaces used to have varying functions. The problem is not intended to be realistic.
mrstModule add ad-core ad-blackoil ad-props mrst-gui
Set up parameters¶
We create a small grid, with initially uniform fluid parameters.
dims = [20, 20];
G = cartGrid(dims, [1, 1]);
G = computeGeometry(G);
fluid = initSimpleADIFluid('phases', 'wo', 'rho', [100, 100],...
'cR', 1e-10/barsa, ...
'mu', [1, 1]);
Computing normals, areas, and centroids... Elapsed time is 0.000118 seconds.
Computing cell volumes and centroids... Elapsed time is 0.000750 seconds.
Add multiple relative permeability functions¶
We create a quadratic and a linear relative-permeability relation as function handles of the saturation. We define the water and oil relative permeability to be equal as cell arrays. In doing so, the first entry corresponds to the first region and the second function to the second region and so on. We also create a cell-wise region indicator to map the two functions we have defined onto the grid. The exact mechanism for how we set up the region in the model can vary.
kr_1 = @(S) S.^2;
kr_2 = @(S) S;
fluid.krW = {kr_1, kr_2};
fluid.krO = {kr_1, kr_2};
rock = makeRock(G, 0.1*darcy, 0.3);
reg = 1 + double(G.cells.centroids(:, 2) > 0.5);
Plot the fluid regions and the corresponding relative permeability curves¶
We have a top and bottom part of the domain
figure;
subplot(1, 2, 1)
plotCellData(G, reg);
axis equal tight
colorbar('horiz')
colormap(lines(2));
subplot(1, 2, 2); hold on;
s = (0:0.05:1)';
nkr = numel(fluid.krW);
l = cell(1, nkr);
for i = 1:nkr
k = fluid.krW{i};
plot(s, k(s), 'linewidth', 2);
l{i} = sprintf('Region %d', i);
end
legend(l, 'location', 'northwest');
xlabel('Saturation');
title('Relative permeability functions')
Set up simulation scenario¶
We inject 1 pore-volume over 100 days, from the left part of the domain to the right.
pv = poreVolume(G, rock);
time = 100*day;
irate = sum(pv)/time;
bc = [];
bc = fluxside(bc, G, 'Xmin', irate, 'sat', [1, 0]);
bc = pside(bc, G, 'XMax', 100*barsa, 'sat', [1, 0]);
state0 = initResSol(G, 100*barsa, [0, 1]);
schedule = simpleSchedule(rampupTimesteps(time, time/10), 'bc', bc);
Approach 1: Use different rock-types¶
We can set the rock.regions.saturation field to set up the different regions for the model. This is the default place the relative permeability and capillary pressure implementations will look for a region.
rock_reg = rock;
rock_reg.regions = struct('saturation', reg);
model = GenericBlackOilModel(G, rock_reg, fluid, 'gas', false);
[~, states] = simulateScheduleAD(state0, model, schedule);
figure;
plotToolbar(G, states, 'startPlayBack', true, 'field', 's:1')
title('Different regions (via rock)');
*****************************************************************
********** Starting simulation: 18 steps, 100 days *********
*****************************************************************
Preparing model for simulation...
Model ready for simulation...
Preparing schedule for simulation...
Schedule ready for simulation...
Validating initial state...
...
Approach 2: Directly modify the state functions¶
For a more fine-grained approach, we can manually update the RelativePermeability state function to have a new region. If a state function is using a function handle from the fluid in it’s implementation and it has a “regions” field set up, it will use that to evaluate the function. Since it is possible to swap out the StateFunctions to change the implementation of a single function, there are many ways to incorporate spatially varying functions.
model = GenericBlackOilModel(G, rock, fluid, 'gas', false);
model = model.validateModel();
model.FlowPropertyFunctions.RelativePermeability.regions = reg;
[~, states2] = simulateScheduleAD(state0, model, schedule);
figure;
plotToolbar(G, states2, 'startPlayBack', true, 'field', 's:1')
title('Different regions (via rock)');
*****************************************************************
********** Starting simulation: 18 steps, 100 days *********
*****************************************************************
Preparing model for simulation...
Model ready for simulation...
Preparing schedule for simulation...
Schedule ready for simulation...
Validating initial state...
...
Approach 3: Use input files¶
We do not demonstrate this in this example, but if you build your model using inputs from the deckformat module, regions are automatically set up if present.
<html>
% <p><font size="-1
Demonstrate Interactive Plotting of Fluid Properties for AD-solvers¶
Generated from fluidInspectionExample.m
The AD-OO framework can interactively visualize the fluid model of a ReservoirModel instance. Once active, the user can interactively explore the different fluid properties (viscosities, relative permeabilities, densities) as functions of saturation and pressure. Load the blackoil module and others to create test data.
mrstModule add ad-blackoil ad-core ad-props deckformat
Inspect the SPE1 fluid model¶
The SPE1 fluid model is a three-phase blackoil model with solution gas. For more information, as well as a simulation example, see the SPE1 blackoil tutorial. We use a setup routine to get a ReservoirModel subclass, and pass it to inspectFluidModel. Note that in this specific case, the tables for undersaturated values can lead to negative or discontinuous values for certain high pressure values. This is not always easy to see directly from the tables, but by using the fluid inspector it is straightforward.
[G, rock, fluid, deck] = setupSPE1();
spe1 = selectModelFromDeck(G, rock, fluid, deck);
inspectFluidModel(spe1, 'pressureRange', (0:10:500)*barsa)
No converter needed in section 'REGIONS'.
No converter needed in section 'SUMMARY'.
Adding 200 artificial cells at top/bottom
Processing regular i-faces
Found 550 new regular faces
Elapsed time is 0.001499 seconds.
...
Inspect the SPE9 fluid model¶
Another standard black-oil test case is the SPE9 model. For more information about this test case, see the SPE9 black-oil tutorial. Of particular interest in this case is the non-zero capillary pressure, and the highly irregular relative permeability curves.
[G, rock, fluid, deck] = setupSPE9();
spe9 = selectModelFromDeck(G, rock, fluid, deck);
inspectFluidModel(spe9)
No converter needed in section 'REGIONS'.
No converter needed in section 'SUMMARY'.
Adding 1200 artificial cells at top/bottom
Processing regular i-faces
Found 10625 new regular faces
Elapsed time is 0.004735 seconds.
...
Set up a two-phase oil-water fluid and inspect it¶
We can use the inspection utility to get a better understanding of how the fluid model changes when we adjust parameters. In this case, we set up a simple two-phase, oil-water model and add in other relative permeability curves with different residual values and Corey exponents. Feel free to modify the parameters and look at how the values change.
fluid = initSimpleADIFluid('phases', 'WO', ...
'rho', [1000, 700], ...
'cR', 1e-8/barsa, ...
'c', [0, 1e-4/barsa]);
srw = 0.2;
sro = 0.3;
% Fluid relative permeabilities (use name convention from SWOF keyword)
fluid.krW = coreyPhaseRelpermAD(2, srw, 1, srw + sro);
fluid.krOW = coreyPhaseRelpermAD(4, sro, 1, srw + sro);
model_ow = TwoPhaseOilWaterModel([], [], fluid);
% Inspect model, and specify pressure range of interest
inspectFluidModel(model_ow, 'pressureRange', (250:10:500)*barsa);
Copyright notice¶
<html>
% <p><font size="-1
Introduction to StateFunctions for the AD-OO framework¶
Generated from stateFunctionTutorial.m
The StateFunction class, together with the StateFunctionGrouping class, is the main mechanism for evaluating properties during a AD-OO simulation. These functions act directly on the “state” object and fully support automatic differentiation, lazy evaluation and different regions.
mrstModule add ad-core ad-blackoil deckformat ad-props
Create a black-oil test model¶
We set up the SPE1 model, a black-oil model with disgas (but no vapoil).
pth = getDatasetPath('spe1');
fn = fullfile(pth, 'BENCH_SPE1.DATA');
[state0, model, schedule, nonlinear] = initEclipseProblemAD(fn);
No converter needed in section 'REGIONS'.
No converter needed in section 'SUMMARY'.
Adding 200 artificial cells at top/bottom
Processing regular i-faces
Found 550 new regular faces
Elapsed time is 0.000880 seconds.
...
Validate model to set up defaults¶
State functions are set up at the start of a simulation, with reasonable defaults given. Here, we manually call validateModel to invoke these defaults.
model = model.validateModel();
% We can now get the groupings belonging to this model
groups = model.getStateFunctionGroupings();
Test the evaluation¶
Each grouping is a collection of one or more StateFunctions, which can depend on each other, or the state of the physical system itself. Dependencies between these are automatically handled during simulations. For instance, let us get the mobility of the initial state. We then plot the oil mobility on the grid.
% We can get this with getProp, just as we would with the pressure or
% saturations which are stored directly in the state. Normally, we do not
% have to think too hard about where exactly a property is defined, since
% the framework can figure it out for us.
mobility = model.getProp(state0, 'Mobility');
% Alternatively, we could invoke the FlowPropertyFunctions grouping's get
% function to the same effect.
mob = model.FlowPropertyFunctions.get(model, state0, 'Mobility');
figure;
plotCellData(model.G, mobility{2});
title('Initial oil mobility')
view(30, 30);
colorbar
We can also initialize AD-variables in state to get derivatives¶
Initialize pressure as a AD-variable
stateAD = state0;
stateAD.pressure = initVariablesADI(state0.pressure);
% Outputs are now ADI, with differentiation with respect to pressure
mobAD = model.FlowPropertyFunctions.get(model, stateAD, 'Mobility');
disp(mobAD)
[1×1 ADI] [1×1 ADI] [1×1 ADI]
Show a state function grouping¶
We can inspect the FlowPropertyFunctions directly. The FlowPropertyFunctions consist of the basic properties required for flow in MRST. Additional properties can be added dynamically to the class instance itself.
disp(model.FlowPropertyFunctions)
<a href="matlab:helpPopup FlowPropertyFunctions">FlowPropertyFunctions</a> (<a href="matlab:edit FlowPropertyFunctions.m">edit</a>) state function grouping instance.
Intrinsic properties (Class properties for FlowPropertyFunctions, always present):
CapillaryPressure: <a href="matlab:helpPopup BlackOilCapillaryPressure">BlackOilCapillaryPressure</a> (<a href="matlab:edit BlackOilCapillaryPressure.m">edit</a>)
ComponentMobility: <a href="matlab:helpPopup ComponentMobility">ComponentMobility</a> (<a href="matlab:edit ComponentMobility.m">edit</a>)
ComponentPhaseDensity: <a href="matlab:helpPopup ComponentPhaseDensity">ComponentPhaseDensity</a> (<a href="matlab:edit ComponentPhaseDensity.m">edit</a>)
ComponentPhaseMass: <a href="matlab:helpPopup ComponentPhaseMass">ComponentPhaseMass</a> (<a href="matlab:edit ComponentPhaseMass.m">edit</a>)
...
Plot dependencies¶
Generally, only a few values are needed to assemble the final linearized equation. Many other functions may be required to get intermediate results, however. For instance, capillary pressure does not occur directly in the flow equations, but standard functions for density, phase pressures and viscosity can depend strongly on the capillary pressure between phases. We can plot the dependency graph of all the state function groupings in order to understand the relationships between the different functions.
for i = 1:numel(groups)
figure;
plotStateFunctionGroupings(groups{i})
title(class(groups{i}));
end
Plot a specific dependency¶
Plot all dependencies for the mobility we just evaluated
figure(1); clf
plotStateFunctionGroupings(model.FlowPropertyFunctions, 'Stop', 'Mobility')
title('All dependencies required to evaluate mobility')
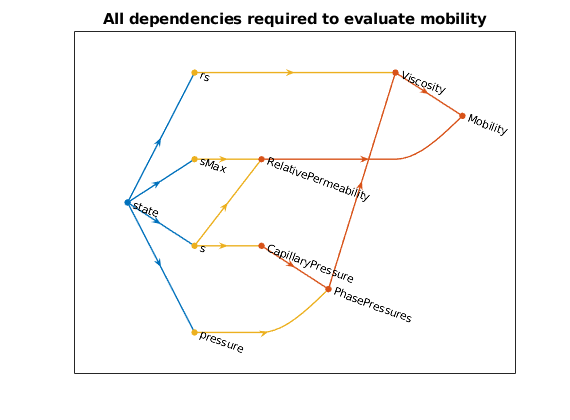
Plot all dependencies of a property¶
Let’s see all properties which directly or indirectly depend on on the pressure in the state
figure(1); clf
plotStateFunctionGroupings(model.FlowPropertyFunctions, 'Start', 'pressure')
title('Flow properties that depend on pressure')
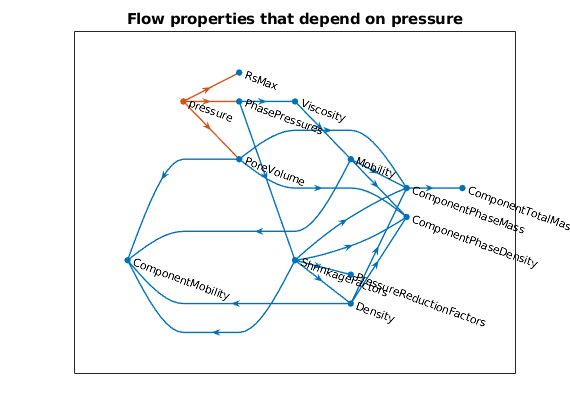
Plot everything which either depends upon a property, or uses that property¶
Let’s see all properties which directly or indirectly depend on on the pressure in the state
figure(1); clf
plotStateFunctionGroupings(model.FlowPropertyFunctions, 'Center', 'Mobility')
title('Upstream and downstream mobility dependencies');
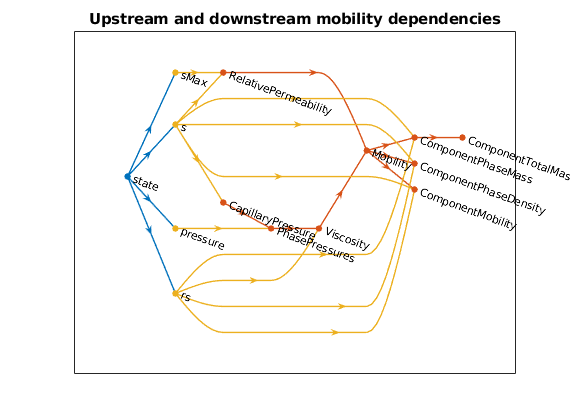
We can combine graphs¶
Here, we plot all the flow properties together with the discretization of the flux, which depends on many of these properties.
df = get(0, 'DefaultFigurePosition');
figure('Position', df.*[1, 1, 2, 2]);
tmp = {model.FlowPropertyFunctions, model.FluxDiscretization};
plotStateFunctionGroupings(tmp);
title('Flow properties + flow discretization')

Create a compositional model and visualize the property graphs¶
We create a compositional model, which has a few major differences from the black-oil model: Note the addition of equation-of-state properties such as fugacity or phase compressibility factors. In addition, the relationship between the ShrinkageFactors and Density
rstModule add compositional
eos = getCompositionalFluidCase('simple');
cmodel = GenericNaturalVariablesModel(model.G, model.rock, model.fluid, eos, 'water', true);
cmodel = cmodel.validateModel();
clf;
plotStateFunctionGroupings(cmodel.FlowPropertyFunctions);
% <html>
% <p><font size="-1
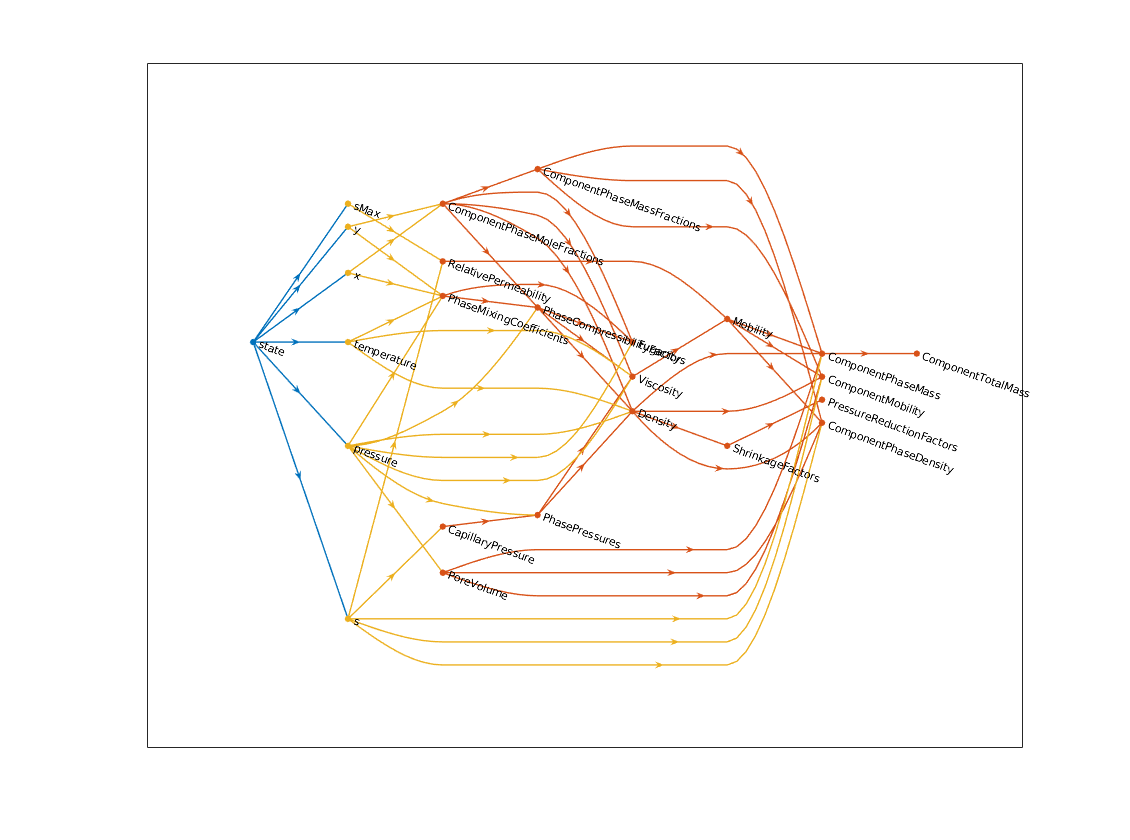
mrstModule add ad-core ad-blackoil ad-props
Set up scenario¶
Generated from wenoExampleAD.m
dims = [50, 50];
pdims = [100, 100];
G = cartGrid(dims, pdims);
G = computeGeometry(G);
rock = makeRock(G, 0.1*darcy, 0.3);
fluid = initSimpleADIFluid('n', [1, 1], 'mu', [1, 1]*centi*poise, 'rho', [100, 100], 'phases', 'wo');
model = GenericBlackOilModel(G, rock, fluid, ...
'water', true, 'oil', true, 'gas', false);
time = 1*year;
irate = 2*sum(model.operators.pv)/time;
W = [];
W = addWell(W, G, rock, 1, 'comp_i', [1, 0], 'val', irate, 'type', 'rate');
W = addWell(W, G, rock, G.cells.num, 'comp_i', [1, 0], 'val', 50*barsa, 'type', 'bhp');
n = 300;
dt = repmat(time/n, n, 1);
schedule = simpleSchedule(dt, 'W', W);
state0 = initResSol(G, 100*barsa, [0, 1]);
Computing normals, areas, and centroids... Elapsed time is 0.001207 seconds.
Computing cell volumes and centroids... Elapsed time is 0.002973 seconds.
Defaulting reference depth to top of formation for well W1. Please specify 'refDepth' optional argument to 'addWell' if you require bhp at a specific depth.
Defaulting reference depth to top of formation for well W2. Please specify 'refDepth' optional argument to 'addWell' if you require bhp at a specific depth.
Simulate base case¶
[ws, states, report] = simulateScheduleAD(state0, model, schedule);
*****************************************************************
********** Starting simulation: 300 steps, 365 days *********
*****************************************************************
Preparing model for simulation...
Model ready for simulation...
Preparing schedule for simulation...
Schedule ready for simulation...
Validating initial state...
...
Set up a WENO discretization¶
model_weno = model;
weno = WENOUpwindDiscretization(model_weno);
Getting supports.
100 of 2500...
200 of 2500...
300 of 2500...
400 of 2500...
500 of 2500...
600 of 2500...
700 of 2500...
...
Override the component discretization with a WENO scheme¶
model_weno = model_weno.validateModel();
props = model_weno.FluxDiscretization;
disp(props)
props = props.setStateFunction('FaceMobility', FaceMobility(model_weno, weno));
props = props.setStateFunction('FaceComponentMobility', FaceComponentMobility(model_weno, weno));
model_weno.FluxDiscretization = props;
[ws_weno, states_weno, report_weno] = simulateScheduleAD(state0, model_weno, schedule);
<a href="matlab:helpPopup FluxDiscretization">FluxDiscretization</a> (<a href="matlab:edit FluxDiscretization.m">edit</a>) state function grouping instance.
Intrinsic properties (Class properties for FluxDiscretization, always present):
ComponentPhaseFlux: <a href="matlab:helpPopup ComponentPhaseFlux">ComponentPhaseFlux</a> (<a href="matlab:edit ComponentPhaseFlux.m">edit</a>)
ComponentTotalFlux: <a href="matlab:helpPopup ComponentTotalFlux">ComponentTotalFlux</a> (<a href="matlab:edit ComponentTotalFlux.m">edit</a>)
FaceComponentMobility: <a href="matlab:helpPopup FaceComponentMobility">FaceComponentMobility</a> (<a href="matlab:edit FaceComponentMobility.m">edit</a>)
GravityPotentialDifference: <a href="matlab:helpPopup GravityPotentialDifference">GravityPotentialDifference</a> (<a href="matlab:edit GravityPotentialDifference.m">edit</a>)
...
model_e = model.validateModel();
fd = model_e.FluxDiscretization;
fd = fd.setFlowStateBuilder(AdaptiveImplicitFlowStateBuilder('initialStep', 0.02*day, 'verbose', true));
model_e.FluxDiscretization = fd;
[ws_e, states_e, report_e] = simulateScheduleAD(state0, model_e, schedule);
*****************************************************************
********** Starting simulation: 300 steps, 365 days *********
*****************************************************************
Preparing model for simulation...
Model ready for simulation...
Preparing schedule for simulation...
Schedule ready for simulation...
Validating initial state...
...
model_weno_expl = model_weno;
fd = model_weno_expl.FluxDiscretization;
fd = fd.setFlowStateBuilder(AdaptiveImplicitFlowStateBuilder('initialStep', 0.02*day, 'verbose', true));
model_weno_expl.FluxDiscretization = fd;
[ws_weno_ex, states_weno_ex, report_weno_ex] = simulateScheduleAD(state0, model_weno_expl, schedule);
*****************************************************************
********** Starting simulation: 300 steps, 365 days *********
*****************************************************************
Preparing model for simulation...
Model ready for simulation...
Preparing schedule for simulation...
Schedule ready for simulation...
Validating initial state...
...
Plot saturations¶
G.cells.sortedCellNodes = getSortedCellNodes(G);
for i = 1:numel(states)
figure(1); clf;
subplot(2, 2, 1)
plotCellData(G, states{i}.s(:, 1), 'edgecolor', 'none');
axis equal tight
title('FIM SPU');
subplot(2, 2, 2)
plotCellData(G, states_weno{i}.s(:, 1), 'edgecolor', 'none');
axis equal tight
title('FIM WENO')
subplot(2, 2, 3)
plotCellData(G, states_e{i}.s(:, 1), 'edgecolor', 'none');
title('AIM SPU')
axis equal tight
subplot(2, 2, 4)
plotCellData(G, states_weno_ex{i}.s(:, 1), 'edgecolor', 'none');
title('AIM WENO')
axis equal tight
end
Plot wells¶
lotWellSols({ws, ws_e, ws_weno, ws_weno_ex}, report.SimulationTime, 'datasetnames', {'FIM-SPU', 'AIM-SPU', 'FIM-WENO', 'AIM-WENO'}, 'field', 'qWs', 'SelectedWells', 2)
% <html>
% <p><font size="-1